Published online Oct 7, 2015. doi: 10.3748/wjg.v21.i37.10563
Peer-review started: April 24, 2015
First decision: June 2, 2015
Revised: June 13, 2015
Accepted: August 25, 2015
Article in press: August 25, 2015
Published online: October 7, 2015
Helicobacter pylori (H. pylori) infection is present in more than half the world’s population and has been associated with several gastric disorders, such as gastritis, peptic ulceration, and gastric adenocarcinoma. The clinical outcome of this infection depends on host and bacterial factors where H. pylori virulence genes seem to play a relevant role. Studies of cagA and vacA genes established that they were determining factors in gastric pathogenesis. However, there are gastric cancer cases that are cagA-negative. Several other virulence genes have been searched for, but these genes remain less well known that cagA and vacA. Thus, this review aimed to establish which genes have been suggested as potentially relevant virulence factors for H. pylori-associated gastrointestinal diseases. We focused on the cag-pathogenicity island, genes with adherence and motility functions, and iceA based on the relevance shown in several studies in the literature.
Core tip:Helicobacter pylori (H. pylori) infection is present in more than half the world’s population and has been associated with several gastric disorders. The clinical outcome of this infection depends on host and bacterial factors. Studies have established that cagA and vacA H. pylori genes are determining factors in gastric pathogenesis. This review aimed to examine which genes have been suggested as potentially relevant virulence factors for H. pylori, focusing on the cag-pathogenicity island, adherence and motility genes, and iceA based on the relevance shown in several studies.
-
Citation: da Costa DM, Pereira EDS, Rabenhorst SHB. What exists beyond
cag A andvac A?Helicobacter pylori genes in gastric diseases. World J Gastroenterol 2015; 21(37): 10563-10572 - URL: https://www.wjgnet.com/1007-9327/full/v21/i37/10563.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i37.10563
Helicobacter pylori (H. pylori) is a spiral shaped Gram-negative bacterium that selectively colonizes the gastric mucous layer by adhering to the epithelial lining of the stomach. It is a urease-, catalase-, and oxidase-positive bacterium that possesses 4 to 6 polar flagella for motility, and several virulence factors which vary with the strain[1,2]. H. pylori was isolated for the first time in 1983 by Warren and Marshall from gastric biopsy samples of patients with chronic gastritis and peptic ulcer. However, the finding of spiral bacteria in the stomach of animals dates back to 1906[3,4].
Evidence suggests that the relationship between H. pylori and its human host has existed for at least 60000 years. One piece of evidence is that the genetic diversity of bacteria evolved in parallel with the human species, showing that both have been evolving intimately ever since. Furthermore, the genetic diversity distribution of H. pylori is consistent with the colonization of the earliest humans and with co-migration out of East Africa[5]. In 1994, the World Health Organization recognized this bacteria as a type I (definite) carcinogen in humans, based on evidence that H. pylori is involved in the development of gastric adenocarcinoma[6]. H. pylori infection is present in more than half the world’s population. However, not all infected people exhibit diseases associated with this bacterium. It is the main cause of gastric disorders, such as gastritis in about 20%, peptic ulceration in 10%, gastric adenocarcinoma in 1%-2%, and gastric MALT lymphoma in less than 0.1% of the people infected[7,8].
The clinical outcome of infection by H. pylori depends on the presence of bacterial virulence factors and on factors related to the host. Several virulence genes have been well studied and established in the literature as determining factors in gastric pathogenesis, such as cagA (cytotoxin-associated gene A) and vacA (vacuolating cytotoxin A) genes. Several other genes, although previously studied, remain less well recognized than cagA and vacA. Thus, the objective of this review is to discuss current knowledge of H. pylori virulence factors that highlight other genes in the cag-pathogenicity island (cag-PAI), genes that code outer membrane proteins (babA, oipA, sabA, hopQ), motility genes (flaA and flaB), and iceA, which have been identified in the literature as potentially relevant in the development of more severe lesions.
The cagA and vacA genes are both well established and extensively studied as H. pylori virulence factors. Whereas not all H. pylori strains possess the cagA gene, essentially, all strains possess the vacA gene. However, not all secrete a VacA product, which depends on the gene structure.
The cagA gene is a recognized marker for the presence of cag-PAI. This gene encodes a 121-145 kDa immuno-dominant protein (CagA) that, when injected into the gastric epithelial cell cytoplasm, interacts with host cell proteins, inducing cell morphological changes (hummingbird phenotype), and pro-inflammatory and mitogenic responses. Several studies in cell culture and animal models indicate the importance of cagA gene involvement in human gastric cancer, and that its deletion prevents the development of the disease in a gerbil model[9-11]. Most of the H. pylori strains in East Asia have the cagA gene, regardless of the disease. Thus, the pathogenic difference in this region is difficult to explain in terms of the presence or absence of the cagA gene alone[12].
The CagA protein contains tyrosine phosphorylation motifs (glutamate-proline-isoleucinetyrosine-alanine, EPIYA) within the carboxyl-terminal variable region of the protein. Studies show the existence of four EPIYA motifs (A, B, C, D). EPIYA-A and EPIYA-B are present throughout the world, EPIYA-C is predominantly found in strains from Western countries, and EPIYA-D is found almost exclusively in East-Asian strains (Japan, South Korea, and China). H. pylori strains containing EPIYA-D motifs induce significantly higher levels of interleukin-8 release from gastric epithelial cells compared with strains containing the A-B-C-type of CagA[13,14].
The vacA gene is not part of the cag-PAI. It induces vacuolization and various cellular activities such as the formation of membrane channels, the release of cytochrome c from mitochondria leading to apoptosis, and binding to cell membrane receptors, followed by a pro-inflammatory response[15,16]. However, considerable differences in vacuolating activities are observed between strains according to variations in the vacA gene structure within the signal (s), middle (m), and intermediate (i) regions[17]. The “s” and “m” regions are stratified into s1 or s2, and m1 or m2 subtypes, and the possible combinations generate proteins with different cytotoxicity. In vitro experiments showed that vacA s1/m1 strains induce greater vacuolation than s1/m2 strains, and there is typically no vacuolating activity in s2/m2 strains[17].
In agreement with in vitro data, studies in the Middle East, Africa, and Western countries have shown that individuals infected with vacA s1 or m1 H. pylori strains have an increased risk of peptic ulcer or gastric cancer compared with individuals infected with s2 or m2 strains[18,19]. On the other hand, in East Asia, as most strains are vacA s1, the differences in pathogenesis cannot be explained by the type of “s” region[20]. In turn, the “m” region in East Asia shows variations suggesting that it may play a role in the regional difference. In northern East Asia, there is a higher prevalence of strains vacA m1 and incidence of gastric cancer, whereas in the south of East Asia, where the strains vacA m2 are prevalent, the incidence of cancer is lower than in the northern region[20,21]. A Brazilian study indicated that in the absence of cagA, there was a role for vacA s1 in the development of gastric cancer, since most of the negative strains had the vacA s1 gene[22].
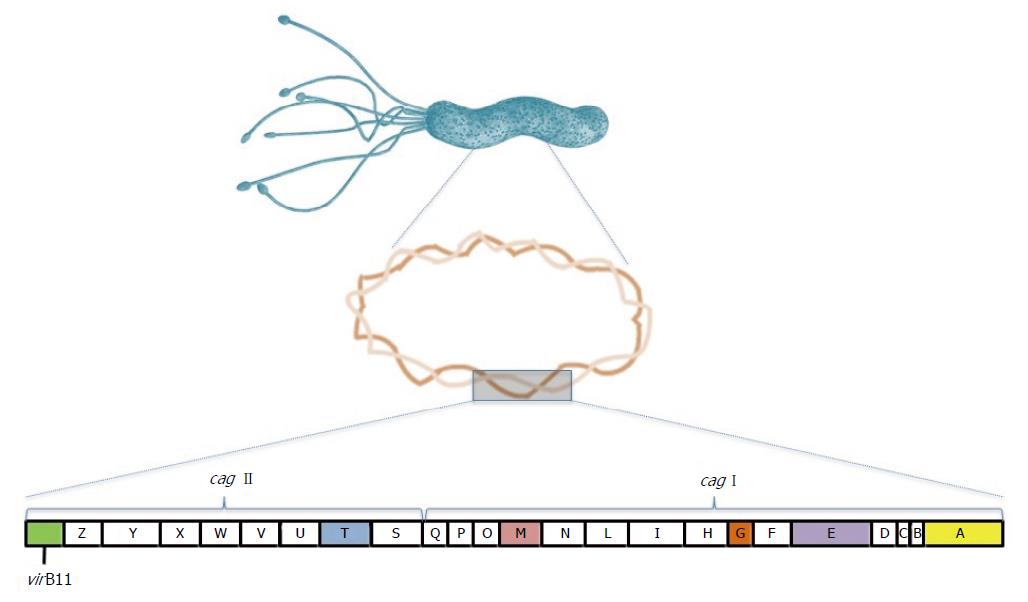
cag-PAI is a segment of H. pylori DNA of 40 kb containing 31 genes[23]. Most of these genes encode functional components of a type 4 bacterial secretion system (T4SS) used for the translocation of bacterial products directly into the host cell cytoplasm, including the cagA gene product[24]. cag-PAI is found in about 60% of Western strains, whereas almost all of the East Asian strains isolated are cag-PAI positive[25]. The positive cag-PAI strains are more related to peptic ulcer and gastric cancer than the negative strains, showing that cag-PAI plays an important role in H. pylori pathogenesis[26,27].
A phylogeny study showed by sequencing that most cag-PAI genes were similar to those of housekeeping genes, indicating that cag-PAI was probably acquired only once by H. pylori. Thus, H. pylori genetic diversity seems to reflect the geographic isolation that has shaped this bacterial species since modern humans migrated out of Africa. Carriage of cag-PAI varies from an almost universal presence in the strains hpEastAsia and hpAfrica1, through an intermediate presence (hpEurope) to complete absence (hpAfrica2). When compared with other bacteria of the same genus, the absence of cag-PAI seems to be an ancestral trait. Thus, the pathogenicity island would have been acquired by horizontal gene transfer from an unknown source after H. pylori had established itself in humans[11].
Initial studies on the integrity of cag-PAI analyzed sequences of cagI and cagII regions and genes present in these regions separately[28,29]. A study that analyzed cag-PAI integrity showed that rearrangement in this island is a prevalent phenomenon, with less rearrangement in the cagE and cagT genes than in the cagA gene. cag-PAI was disrupted in the majority of isolated strains throughout the world. Conservation of cag-PAI was highest in Japanese isolates and minimal in European and African strains[30]. Infection with a strain containing a complete set of cag-PAI genes was associated with the development of ulcer disease, showing the importance of these genes to gastric diseases outcomes[31].
Several Cag proteins have been detected as constituents of H. pylori cag T4SS apparatus and have important roles in the translocation of CagA[32]. These include CagE, VirB11, CagT, CagM and CagG, whose importance will be described below (Figure 1).
cagE is located in the right half of cag-PAI, and studies have suggested that this gene is a more accurate marker of an intact pathogenicity island and can be used as a cag-PAI marker with cagA[33,34]. virB11 codifies a protein that has a ring-shaped structure composed of 6 monomeric units. These genes code transmembrane proteins with ATPase activity that provides the energy for apparatus assembly and/or substrate transport[24,35].
Although there is a well-established relationship between cagE and virB11 genes with gastritis, peptic ulcer, and duodenal ulcer, only a few studies have described an association with gastric cancer[36-38]. Two of these studies showed the presence of such genes in early tumor stages and an association with other virulence genes, indicating that there is a role for cagE and virB11 in gastric carcinogenesis[34,39].
The cagT gene is a homologue of A. tumefaciens vir B7 and has been reported to be a marker of the cagII region[40]. Some studies revealed that CagT localized in both inner and outer membranes plays an important role in the induction of the proinflammatory cytokine interleukin-8 when localized in the outer membrane[41-43]. It may also interact with CagA and facilitate its translocation into host cells, acting as a chaperone-like protein localized in the inner membrane[44].
The expression of CagT in H. pylori is also closely associated with severe gastric disease[45]. Deletion frequencies of cagT genes were higher in benign cases compared with isolates from severe ulcers and gastric cancers[46,47]. Studies reported an association of the cagT gene with the development of peptic ulcer disease, suggesting a high virulence gene in H. pylori[40,48,49]. The cagT gene, as well as the cagA gene, have been associated with other virulence factors, such as vacA s1, vacA m1, and the genotype vacA s1m1, occurring in smaller percentages concomitantly with vacA s2 strains, vacA m2, and vacA s2m2[49,50]. Therefore, CagT seems to be a very important protein in H. pylori, not only for the integrity of the cag-PAI apparatus, but also for determining disease severity.
The cagM gene has been reported to be a marker of the cagI region[51]. Some studies revealed that the protein encoded by this gene forms a surface structure which acts as a nuclear factor (NF)-κB-inducing agent, mediating interleukin-8 secretion[41,52,53]. It is also involved in the repression of H/K-ATPase transcription, which causes the downregulation of human gastric H/K-ATPase expression, significantly inhibiting acid secretion by gastric cells[54,55]. CagM expression may represent a first line of H. pylori defense against gastric acid, which may otherwise be upregulated by the presence of CagM-deficient Gram-negative bacteria.
Expression of CagM in H. pylori is also associated with severe gastric disease. Some studies revealed that the cagM gene was associated with the development of gastritis, peptic ulcers, and gastric cancer[45,49,56,57]. Thus, CagM is a very important protein, not only for the integrity of the cag-PAI apparatus, but also for determining disease severity, and a line of H. pylori defense against gastric acid.
cagG is located in the right side of cag-PAI, and it has been reported to be a marker of the cagI region. This gene is not a vir homologue, but it has weak homology with the flagellar motor switch protein gene or toxin co-regulated pilus biosynthesis protein gene[23,51,58]. It may also play an important role in the induction of the proinflammatory cytokine interleukin-8[41,52].
Some studies suggest that cagG may have a function related to adhesion to gastric epithelial cells. cagG-deleted strains adhere less to gastric epithelial cells, and these strains cause a reduction in the amount of interleukin-8 secreted from the cells[59,60]. The frequency of the cagG gene has been high in several gastrointestinal diseases, but a specific disease related to it has not been established[51,61].
Given the above, we suggest that the integrity of cag-PAI seems to be more relevant than the presence of the gene alone. It is believed that the presence of cagA alone is not sufficient for bacterial pathogenicity, but the set of genes which form an efficient T4SS confers pathogenicity.
Approximately 4% of the H. pylori genome encodes a diverse repertoire of Outer membrane proteins (OMPs) that have been grouped into 5 major families[62]. The Helicobacter outer membrane protein (Hop) family is the largest and includes adhesins such as BabA (HopS), SabA (HopP), OipA (HopH) and HopQ. Adherence of H. pylori to the gastric mucosa plays important roles in the initial colonization and long-term persistence on the gastric mucosa, as well as in the intensity of the resulting inflammatory response[63].
Blood group antigen-binding adhesin (babA) is a 78-kDa outer membrane protein encoded by the babA2 gene, which binds the fucosylated Lewisb antigen (Leb) on the surfaces of gastric epithelial cells and is the best described H. pylori OMP[64,65]. Although three bab alleles have been identified (babA1, babA2, and babB), only the babA2 gene product is functionally active[66]. Analyses of binding characteristics of H. pylori strains worldwide suggest that BabA has evolved in response to host mucosal glycosylation patterns to permit H. pylori to adapt to its host and to maintain persistent colonization[67].
Some researchers have demonstrated that babA2 is associated with increased risk of duodenal ulcer disease and adenocarcinoma, and when found in conjunction with the cagA and vacA s1 alleles, leads to an even greater risk of developing more severe diseases[68,69]. BabA binding to Leb is also important for the induction of DNA double-strand breaks in host cell lines, and may promote cancer-associated gene mutations[70]. Adherence via BabA also enhances the ability of the type IV secretion apparatus to contact host cells, leading to a stronger inflammatory response[71]. Therefore, BabA is important not only for H. pylori to adhere to the stomach surface but also to anchor the bacterial secretion system to the host cell surface so that bacterial factors can be effectively injected into the host cell cytosol.
The sialic acid-binding adhesin, SabA or HopP or OMP17 (about 70 KDa) is the second best characterized adhesin of H. pylori, and binds sialyl-Lewis antigens that are expressed on inflamed gastric tissue[64,72]. H. pylori modulates the expression of the SabA ligand, the sialyl-dimeric-Lex, in human gastric cell lines via the induction of a specific glycosyltransferase, β3 GlcNAc T5 (β3GnT5), involved in the biosynthesis of Lewis antigens, thereby strengthening the epithelial attachment necessary to achieve successful colonization[73].
The sabA gene expression is regulated at transcriptional level by some mechanisms. Indeed, the dinucleotide CT repeats present in the 5’ coding region of sabA regulates their expression by phase variation through a slipped strand repair mechanism (SSM)[74,75], and the sabA promoter region modulates its transcriptional activity through a variable homopolymeric thymidine tract[76]. The frequent “on/off” switch of SabA expression suggests that SabA expression can rapidly respond to changes exerted by the gastric niche. SabA-positive status is inversely related to the ability of the stomach to secrete acid, suggesting that its expression may be regulated by changes in acid secretion and/or in antigens expressed by the atrophic mucosa[67,75].
SabA-positive status is associated with the development of intestinal metaplasia, gastric atrophy and gastric cancer[65,68]. After H. pylori induces gastritis, neutrophils and monocytes infiltrate into the gastric mucosa. SabA of non-opsonized H. pylori strains specifically binds to neutrophils through sialylated carbohydrates. Consequently, the stimulated neutrophils produce reactive oxygen species causing oxidative damage of the gastric epithelium, showing that SabA is a virulence factor[72,77].
OipA (about 34 kDa) was identified in 2000. It is one of the OMPs. It functions in adhesion and is located approximately 100 kbp from the cag-PAI on the H. pylori chromosome[58,78,79]. The functional status of OipA is regulated by slipped strand mispairing that is determined by the number of CT dinucleotide repeated in the 5′ region of the gene (switch “on” and OipA is functional; switch “off” and OipA is nonfunctional)[80].
H. pylori with the OipA functional status “on” has been associated with other virulence factors, such as cag PAI, vacA, iceA and babA[65,68,81,82]. OipA “on” status is significantly associated with more severe gastric diseases (duodenal ulcer and gastric cancer), high H. pylori density, severe neutrophil infiltration, and high mucosal interleukin-8 levels[83]. Researchers have demonstrated that OipA can induce inflammation and affect actin dynamics through the phosphorylation of multiple signaling pathways that usually interact with cag-PAI (CagA)-related pathways[84,85]. H. pylori-related inflammatory signaling related to gastric carcinogenesis is regulated by the activation of the phosphoinositide-3 kinase (PI3K)/Akt signaling pathway[86]. OipA regulates IL-8 secretion through PI3K/Akt and this regulation is dependent on forkhead transcription factors of class O (FoxO) 1/3a inactivation[87]. Inactivation of oipA also results in a decreased level of nuclear β-catenin in vitro and a reduced incidence of cancer in gerbils, indicative of this OMP’s importance in H. pylori virulence[10].
The hopQ gene encodes HopQ, an OMP that attenuates the adherence of H. pylori strains to gastric epithelial cells and thus may play an important role in the initial colonization and long-term persistence of the bacterium in the stomach[88]. The hopQ gene is present in 2 forms: types I and II. Some studies have reported an association between the presence of type I hopQ alleles and other H. pylori virulence markers, including type s1 vacA alleles[89-92]. In Western patients, the inflammatory cell infiltration and atrophy scores were significantly higher in patients with hopQ type I strains than those with type II[63]. Only one study so far showed that the hopQII genotype is frequently present in H. pylori strains isolated from gastric cancer patients[93].
Another study conducted an analysis of 3000 H. pylori mutants and revealed that the hopQ gene affected NF-κB nuclear translocation. HopQ was essential for CagA translocation and for CagA-mediated host cell responses such as formation of the hummingbird phenotype and cell scattering. It also showed that the deletion of hopQ reduced T4SS-dependent activation of NF-κB, induction of MAPK signaling and secretion of interleukin-8 in the host cells, but it did not affect motility or the quantity of bacteria attached to host cells. Therefore, HopQ exhibits adhesive properties and could be useful in facilitating contact of H. pylori’s T4SS with the host cell surface[94].
Although BabA and SabA are the most prominent adhesins described so far, it seems probable that additional adhesins described in this review are involved in the colonization process. The adhesins are important not only for H. pylori to adhere to the stomach surface but also to anchor the bacterial secretion system and consequently the delivery of virulence factors to host epithelial cells.
Flagella provide the motility of H. pylori which possess a unipolar bundle of 3 to 5 flagella, composed of 3 structural elements: the basal body, the hook, and the filament[67,95,96]. The filament acts as a propeller when rotated at its base and it is composed of 2 flagellins: the major, FlaA, and the minor, FlaB[97]. Mutation of flaA results in flagellar truncation and decreased motility in vitro[98]. In vivo, FlaA and other proteins necessary for flagellar assembly are essential for persistent infection in rodent and gnotobiotic piglet models[99-101].
H. pylori flagellin filaments are post-translationally modified by glycosylation with a 9-carbon pseudaminic acid (Pse) sugar derivative that resembles sialic acid, which is typically found on mammalian cell surfaces[102]. The FlaA protein is modified with a total of 7 O-linked pseudaminic acid (Pse5Ac7Ac) residues, while FlaB is modified with 10 O-linked Pse5Ac7Ac residues. Deletion of genes responsible for the glycosylation process leads to loss of late flagellar structures (hook and filaments) and loss of motility[103,104]. Motility is essential for successful gastric colonization and may contribute to pathogenesis.
Another virulence gene designated iceA (induced by contact with epithelium) has been recently described. Some studies showed that iceA has two main allelic variants, iceA1 and iceA2, but the function of these variants is not yet clear[105,106]. iceA1 demonstrated sequence homology with a gene from Neisseria lactamica, nlaIIIR, which encodes a CTAG-specific restriction endonuclease[107]. On the other hand, iceA2 has no homology to known genes and the function of the iceA2 product remains unclear. The expression of iceA1 is upregulated on contact between H. pylori and human epithelial cells, and the iceA1 genotype was linked with enhanced mucosal interleukin-8 expression and acute antral inflammation[61].
Some reports have reported an association between iceA allelic types and clinical outcomes[108]. The iceA1 variant was associated with peptic ulcer disease, and iceA2 variants with gastritis[109,110]. However, these associations vary among populations. In Brazil, for instance, the iceA1 allele is associated with gastritis[111]. Additionally, in Cuba, Europe, Saudi Arabia, and Turkey the iceA2 allele is associated with non-peptic ulcer dyspepsia as well as strains with more virulent types[109,112]. Thus, the iceA gene may be considered a useful marker in patients with gastroduodenal diseases.
The relationship between H. pylori and humans dates back 50000 years and during this time these 2 species have co-evolved. During this evolution, there has been a major change in the genome of the bacterium with horizontal acquisition of the cag pathogenicity island, which seems to be important in colonization, and responsible for the development of gastric diseases. Although only the cagA gene is well defined as an H. pylori pathogenicity marker, over the course of our review it was observed that other genes are also essential components for a functional cag T4SS. Furthermore, the fact that some strains with an incomplete pathogenicity island in more severe gastric lesions were observed, suggests that there must be genes with overlapping roles to ensure the functioning of the secretory apparatus. In addition, the product of some of these genes could be capable of stimulating an exacerbated inflammatory response which is characteristic of gastric lesions. Although there are several genes associated with adhesion of the bacteria, the babA gene is associated with successful colonization.
P- Reviewer: Carri JH, Vermi W S- Editor: Ma YJ L- Editor: Cant MR E- Editor: Wang CH
| 1. | Goodwin CS, Armstrong JA. Microbiological aspects of Helicobacter pylori (Campylobacter pylori). Eur J Clin Microbiol Infect Dis. 1990;9:1-13. [PubMed] [Cited in This Article: ] |
| 2. | Wen S, Moss SF. Helicobacter pylori virulence factors in gastric carcinogenesis. Cancer Lett. 2009;282:1-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 104] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 3. | Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311-1315. [PubMed] [Cited in This Article: ] |
| 4. | Siqueira JS, Lima PSS, Barreto AS, Quintans Jr L. Aspectos gerais nas infecções por Helicobacter pylori: Revisão. Rev Bras Anal Clin. 2007;39:9-13. [Cited in This Article: ] |
| 5. | Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915-918. [PubMed] [Cited in This Article: ] |
| 6. | Schistosomes , liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1-241. [PubMed] [Cited in This Article: ] |
| 7. | Peek RM, Crabtree JE. Helicobacter infection and gastric neoplasia. J Pathol. 2006;208:233-248. [PubMed] [Cited in This Article: ] |
| 8. | Oluwasola AO. Genetic determinants and clinico-pathological outcomes of helicobacter pylori infection. Ann Ib Postgrad Med. 2014;12:22-30. [PubMed] [Cited in This Article: ] |
| 9. | Murata-Kamiya N, Kurashima Y, Teishikata Y, Yamahashi Y, Saito Y, Higashi H, Aburatani H, Akiyama T, Peek RM, Azuma T. Helicobacter pylori CagA interacts with E-cadherin and deregulates the beta-catenin signal that promotes intestinal transdifferentiation in gastric epithelial cells. Oncogene. 2007;26:4617-4626. [PubMed] [Cited in This Article: ] |
| 10. | Franco AT, Johnston E, Krishna U, Yamaoka Y, Israel DA, Nagy TA, Wroblewski LE, Piazuelo MB, Correa P, Peek RM. Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 2008;68:379-387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 224] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 11. | Olbermann P, Josenhans C, Moodley Y, Uhr M, Stamer C, Vauterin M, Suerbaum S, Achtman M, Linz B. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6:e1001069. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 164] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 12. | Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7:629-641. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 420] [Cited by in F6Publishing: 428] [Article Influence: 30.6] [Reference Citation Analysis (1)] |
| 13. | Argent RH, Hale JL, El-Omar EM, Atherton JC. Differences in Helicobacter pylori CagA tyrosine phosphorylation motif patterns between western and East Asian strains, and influences on interleukin-8 secretion. J Med Microbiol. 2008;57:1062-1067. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 74] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 14. | Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: The master key hypothesis. Helicobacter. 2010;15:163-176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 170] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 15. | Cover TL, Blanke SR. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat Rev Microbiol. 2005;3:320-332. [PubMed] [Cited in This Article: ] |
| 16. | Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449-490. [PubMed] [Cited in This Article: ] |
| 17. | Atherton JC, Cao P, Peek RM, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771-17777. [PubMed] [Cited in This Article: ] |
| 18. | Sugimoto M, Zali MR, Yamaoka Y. The association of vacA genotypes and Helicobacter pylori-related gastroduodenal diseases in the Middle East. Eur J Clin Microbiol Infect Dis. 2009;28:1227-1236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 19. | Sugimoto M, Yamaoka Y. The association of vacA genotype and Helicobacter pylori-related disease in Latin American and African populations. Clin Microbiol Infect. 2009;15:835-842. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 76] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 20. | Yamaoka Y, Orito E, Mizokami M, Gutierrez O, Saitou N, Kodama T, Osato MS, Kim JG, Ramirez FC, Mahachai V. Helicobacter pylori in North and South America before Columbus. FEBS Lett. 2002;517:180-184. [PubMed] [Cited in This Article: ] |
| 21. | Uchida T, Nguyen LT, Takayama A, Okimoto T, Kodama M, Murakami K, Matsuhisa T, Trinh TD, Ta L, Ho DQ. Analysis of virulence factors of Helicobacter pylori isolated from a Vietnamese population. BMC Microbiol. 2009;9:175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 22. | Do Carmo AP, Rabenhorst SH. Importance of vacAs1 gene in gastric cancer patients infected with cagA-negative Helicobacter pylori. APMIS. 2011;119:485-486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 23. | Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648-14653. [PubMed] [Cited in This Article: ] |
| 24. | Terradot L, Waksman G. Architecture of the Helicobacter pylori Cag-type IV secretion system. FEBS J. 2011;278:1213-1222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 25. | Sahara S, Sugimoto M, Vilaichone RK, Mahachai V, Miyajima H, Furuta T, Yamaoka Y. Role of Helicobacter pylori cagA EPIYA motif and vacA genotypes for the development of gastrointestinal diseases in Southeast Asian countries: a meta-analysis. BMC Infect Dis. 2012;12:223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 26. | Nomura AM, Pérez-Pérez GI, Lee J, Stemmermann G, Blaser MJ. Relation between Helicobacter pylori cagA status and risk of peptic ulcer disease. Am J Epidemiol. 2002;155:1054-1059. [PubMed] [Cited in This Article: ] |
| 27. | Matos JI, de Sousa HA, Marcos-Pinto R, Dinis-Ribeiro M. Helicobacter pylori CagA and VacA genotypes and gastric phenotype: a meta-analysis. Eur J Gastroenterol Hepatol. 2013;25:1431-1441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 28. | Audibert C, Burucoa C, Janvier B, Fauchère JL. implication of the structure of the Helicobacter pylori cag pathogenicity island in induction of interleukin-8 secretion. Infect Immun. 2001;69:1625-1629. [PubMed] [Cited in This Article: ] |
| 29. | Owen RJ, Peters TM, Varea R, Teare EL, Saverymuttu S. Molecular epidemiology of Helicobacter pylori in England: prevalence of cag pathogenicity island markers and IS605 presence in relation to patient age and severity of gastric disease. FEMS Immunol Med Microbiol. 2001;30:65-71. [PubMed] [Cited in This Article: ] |
| 30. | Kauser F, Khan AA, Hussain MA, Carroll IM, Ahmad N, Tiwari S, Shouche Y, Das B, Alam M, Ali SM. The cag pathogenicity island of Helicobacter pylori is disrupted in the majority of patient isolates from different human populations. J Clin Microbiol. 2004;42:5302-5308. [PubMed] [Cited in This Article: ] |
| 31. | Talarico S, Gold BD, Fero J, Thompson DT, Guarner J, Czinn S, Salama NR. Pediatric Helicobacter pylori isolates display distinct gene coding capacities and virulence gene marker profiles. J Clin Microbiol. 2009;47:1680-1688. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 32. | Johnson EM, Gaddy JA, Voss BJ, Hennig EE, Cover TL. Genes required for assembly of pili associated with the Helicobacter pylori cag type IV secretion system. Infect Immun. 2014;82:3457-3470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 33. | Sozzi M, Tomasini ML, Vindigni C, Zanussi S, Tedeschi R, Basaglia G, Figura N, De Paoli P. Heterogeneity of cag genotypes and clinical outcome of Helicobacter pylori infection. J Lab Clin Med. 2005;146:262-270. [PubMed] [Cited in This Article: ] |
| 34. | Lima VP, Silva-Fernandes IJ, Alves MK, Rabenhorst SH. Prevalence of Helicobacter pylori genotypes (vacA, cagA, cagE and virB11) in gastric cancer in Brazilian’s patients: an association with histopathological parameters. Cancer Epidemiol. 2011;35:e32-e37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 35. | Kutter S, Buhrdorf R, Haas J, Schneider-Brachert W, Haas R, Fischer W. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J Bacteriol. 2008;190:2161-2171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 96] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 36. | Chomvarin C, Namwat W, Chaicumpar K, Mairiang P, Sangchan A, Sripa B, Tor-Udom S, Vilaichone RK. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA and babA2 genotypes in Thai dyspeptic patients. Int J Infect Dis. 2008;12:30-36. [PubMed] [Cited in This Article: ] |
| 37. | Ramis IB, Vianna JS, Silva Junior LV, Von Groll A, Silva PE. cagE as a biomarker of the pathogenicity of Helicobacter pylori. Rev Soc Bras Med Trop. 2013;46:185-189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 38. | de Negreiros-Bessa P, Barbosa F, do Carmo A, Furtado G, Barroso F, Rabenhorst S. Presence of the Genes cagA, cagE, virB11 and Allelic Variation of vacA of Helicobacter pylori Are Associated with the Activity of Gastritis. Open J Gastroenterol. 2014;4:347-355. [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 39. | Lima VP, de Lima MA, Ferreira MV, Barros MA, Rabenhorst SH. The relationship between Helicobacter pylori genes cagE and virB11 and gastric cancer. Int J Infect Dis. 2010;14:e613-e617. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 40. | Mattar R, Marques SB, Monteiro Mdo S, Dos Santos AF, Iriya K, Carrilho FJ. Helicobacter pylori cag pathogenicity island genes: clinical relevance for peptic ulcer disease development in Brazil. J Med Microbiol. 2007;56:9-14. [PubMed] [Cited in This Article: ] |
| 41. | Fischer W, Püls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42:1337-1348. [PubMed] [Cited in This Article: ] |
| 42. | Rohde M, Püls J, Buhrdorf R, Fischer W, Haas R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol. 2003;49:219-234. [PubMed] [Cited in This Article: ] |
| 43. | Tanaka J, Suzuki T, Mimuro H, Sasakawa C. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol. 2003;5:395-404. [PubMed] [Cited in This Article: ] |
| 44. | Ding H, Zeng H, Huang L, Dong Y, Duan Y, Mao X, Guo G, Zou Q. Helicobacter pylori chaperone-like protein CagT plays an essential role in the translocation of CagA into host cells. J Microbiol Biotechnol. 2012;22:1343-1349. [PubMed] [Cited in This Article: ] |
| 45. | Lai CH, Perng CL, Lan KH, Lin HJ. Association of IS605 and cag-PAI of Helicobacter pylori Isolated from Patients with Gastrointestinal Diseases in Taiwan. Gastroenterol Res Pract. 2013;2013:356217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 46. | Ikenoue T, Maeda S, Ogura K, Akanuma M, Mitsuno Y, Imai Y, Yoshida H, Shiratori Y, Omata M. Determination of Helicobacter pylori virulence by simple gene analysis of the cag pathogenicity island. Clin Diagn Lab Immunol. 2001;8:181-186. [PubMed] [Cited in This Article: ] |
| 47. | Kauser F, Hussain MA, Ahmed I, Srinivas S, Devi SM, Majeed AA, Rao KR, Khan AA, Sechi LA, Ahmed N. Comparative genomics of Helicobacter pylori isolates recovered from ulcer disease patients in England. BMC Microbiol. 2005;5:32. [PubMed] [Cited in This Article: ] |
| 48. | Tiwari SK, Khan AA, Ahmed KS, Ali SM, Ahmed I, Habeeb A, Kauser F, Hussain MA, Ahmed N, Habibullah CM. Polymerase chain reaction based analysis of the cytotoxin associated gene pathogenicity island of Helicobacter pylori from saliva: an approach for rapid molecular genotyping in relation to disease status. J Gastroenterol Hepatol. 2005;20:1560-1566. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 212] [Cited by in F6Publishing: 230] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 49. | Pacheco AR, Proença-Módena JL, Sales AI, Fukuhara Y, da Silveira WD, Pimenta-Módena JL, de Oliveira RB, Brocchi M. Involvement of the Helicobacter pylori plasticity region and cag pathogenicity island genes in the development of gastroduodenal diseases. Eur J Clin Microbiol Infect Dis. 2008;27:1053-1059. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 50. | Roesler BM. Detection and analysis of different genotypes of Helicobacter pylori strains obtained from patients with early and advanced distal type intestinal gastric adenocarcinoma. Accessed on 19 April, 2015. Available from: http://www.bibliotecadigital.unicamp.br/document/?code=000801865. [Cited in This Article: ] |
| 51. | Hsu PI, Hwang IR, Cittelly D, Lai KH, El-Zimaity HM, Gutierrez O, Kim JG, Osato MS, Graham DY, Yamaoka Y. Clinical presentation in relation to diversity within the Helicobacter pylori cag pathogenicity island. Am J Gastroenterol. 2002;97:2231-2238. [PubMed] [Cited in This Article: ] |
| 52. | Glocker E, Lange C, Covacci A, Bereswill S, Kist M, Pahl HL. Proteins encoded by the cag pathogenicity island of Helicobacter pylori are required for NF-kappaB activation. Infect Immun. 1998;66:2346-2348. [PubMed] [Cited in This Article: ] |
| 53. | Smolka AJ, Backert S. How Helicobacter pylori infection controls gastric acid secretion. J Gastroenterol. 2012;47:609-618. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 60] [Article Influence: 5.0] [Reference Citation Analysis (1)] |
| 54. | Saha A, Hammond CE, Beeson C, Peek RM, Smolka AJ. Helicobacter pylori represses proton pump expression and inhibits acid secretion in human gastric mucosa. Gut. 2010;59:874-881. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 55. | Ling F, Wang X, Dai D, Yu M, Chen C, Qian J, Liu C, Zhang Y, Ding J, Guan XW. The Helicobacter pylori protein CagM is located in the transmembrane channel that is required for CagA translocation. Curr Microbiol. 2013;67:531-536. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 56. | Kidd M, Lastovica AJ, Atherton JC, Louw JA. Conservation of the cag pathogenicity island is associated with vacA alleles and gastroduodenal disease in South African Helicobacter pylori isolates. Gut. 2001;49:11-17. [PubMed] [Cited in This Article: ] |
| 57. | Wang MY, Chen C, Gao XZ, Li J, Yue J, Ling F, Wang XC, Shao SH. Distribution of Helicobacter pylori virulence markers in patients with gastroduodenal diseases in a region at high risk of gastric cancer. Microb Pathog. 2013;59-60:13-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 58. | Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539-547. [PubMed] [Cited in This Article: ] |
| 59. | Mizushima T, Sugiyama T, Kobayashi T, Komatsu Y, Ishizuka J, Kato M, Asaka M. Decreased adherence of cagG-deleted Helicobacter pylori to gastric epithelial cells in Japanese clinical isolates. Helicobacter. 2002;7:22-29. [PubMed] [Cited in This Article: ] |
| 60. | Saito H, Yamaoka Y, Ishizone S, Maruta F, Sugiyama A, Graham DY, Yamauchi K, Ota H, Miyagawa S. Roles of virD4 and cagG genes in the cag pathogenicity island of Helicobacter pylori using a Mongolian gerbil model. Gut. 2005;54:584-590. [PubMed] [Cited in This Article: ] |
| 61. | Xu C, Li ZS, Tu ZX, Xu GM, Gong YF, Man XH. Distribution of cagG gene in Helicobacter pylori isolates from Chinese patients with different gastroduodenal diseases and its clinical and pathological significance. World J Gastroenterol. 2003;9:2258-2260. [PubMed] [Cited in This Article: ] |
| 62. | Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun. 2000;68:4155-4168. [PubMed] [Cited in This Article: ] |
| 63. | Ohno T, Sugimoto M, Nagashima A, Ogiwara H, Vilaichone RK, Mahachai V, Graham DY, Yamaoka Y. Relationship between Helicobacter pylori hopQ genotype and clinical outcome in Asian and Western populations. J Gastroenterol Hepatol. 2009;24:462-468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 64. | Borén T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892-1895. [PubMed] [Cited in This Article: ] |
| 65. | Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Borén T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373-377. [PubMed] [Cited in This Article: ] |
| 66. | Pride DT, Meinersmann RJ, Blaser MJ. Allelic Variation within Helicobacter pylori babA and babB. Infect Immun. 2001;69:1160-1171. [PubMed] [Cited in This Article: ] |
| 67. | Wroblewski LE, Peek RM, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713-739. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 817] [Cited by in F6Publishing: 886] [Article Influence: 63.3] [Reference Citation Analysis (1)] |
| 68. | Gerhard M, Lehn N, Neumayer N, Borén T, Rad R, Schepp W, Miehlke S, Classen M, Prinz C. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci USA. 1999;96:12778-12783. [PubMed] [Cited in This Article: ] |
| 69. | Torres LE, Melián K, Moreno A, Alonso J, Sabatier CA, Hernández M, Bermúdez L, Rodríguez BL. Prevalence of vacA, cagA and babA2 genes in Cuban Helicobacter pylori isolates. World J Gastroenterol. 2009;15:204-210. [PubMed] [Cited in This Article: ] |
| 70. | Toller IM, Neelsen KJ, Steger M, Hartung ML, Hottiger MO, Stucki M, Kalali B, Gerhard M, Sartori AA, Lopes M. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc Natl Acad Sci USA. 2011;108:14944-14949. [PubMed] [Cited in This Article: ] |
| 71. | Ishijima N, Suzuki M, Ashida H, Ichikawa Y, Kanegae Y, Saito I, Borén T, Haas R, Sasakawa C, Mimuro H. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J Biol Chem. 2011;286:25256-25264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 137] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 72. | Oleastro M, Ménard A. The Role of Helicobacter pylori Outer Membrane Proteins in Adherence and Pathogenesis. Biology (Basel). 2013;2:1110-1134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 100] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 73. | Marcos NT, Magalhães A, Ferreira B, Oliveira MJ, Carvalho AS, Mendes N, Gilmartin T, Head SR, Figueiredo C, David L. Helicobacter pylori induces beta3GnT5 in human gastric cell lines, modulating expression of the SabA ligand sialyl-Lewis x. J Clin Invest. 2008;118:2325-2336. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 56] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 74. | Yamaoka Y, Kita M, Kodama T, Imamura S, Ohno T, Sawai N, Ishimaru A, Imanishi J, Graham DY. Helicobacter pylori infection in mice: Role of outer membrane proteins in colonization and inflammation. Gastroenterology. 2002;123:1992-2004. [PubMed] [Cited in This Article: ] |
| 75. | Yamaoka Y, Ojo O, Fujimoto S, Odenbreit S, Haas R, Gutierrez O, El-Zimaity HM, Reddy R, Arnqvist A, Graham DY. Helicobacter pylori outer membrane proteins and gastroduodenal disease. Gut. 2006;55:775-781. [PubMed] [Cited in This Article: ] |
| 76. | Kao CY, Sheu BS, Sheu SM, Yang HB, Chang WL, Cheng HC, Wu JJ. Higher motility enhances bacterial density and inflammatory response in dyspeptic patients infected with Helicobacter pylori. Helicobacter. 2012;17:411-416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 77. | Unemo M, Aspholm-Hurtig M, Ilver D, Bergström J, Borén T, Danielsson D, Teneberg S. The sialic acid binding SabA adhesin of Helicobacter pylori is essential for nonopsonic activation of human neutrophils. J Biol Chem. 2005;280:15390-15397. [PubMed] [Cited in This Article: ] |
| 78. | Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27:3325-3333. [PubMed] [Cited in This Article: ] |
| 79. | Yamaoka Y, Kwon DH, Graham DY. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc Natl Acad Sci USA. 2000;97:7533-7538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 305] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 80. | Saunders NJ, Peden JF, Hood DW, Moxon ER. Simple sequence repeats in the Helicobacter pylori genome. Mol Microbiol. 1998;27:1091-1098. [PubMed] [Cited in This Article: ] |
| 81. | Ando T, Peek RM, Pride D, Levine SM, Takata T, Lee YC, Kusugami K, van der Ende A, Kuipers EJ, Kusters JG. Polymorphisms of Helicobacter pylori HP0638 reflect geographic origin and correlate with cagA status. J Clin Microbiol. 2002;40:239-246. [PubMed] [Cited in This Article: ] |
| 82. | Yamaoka Y, Kikuchi S, el-Zimaity HM, Gutierrez O, Osato MS, Graham DY. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology. 2002;123:414-424. [PubMed] [Cited in This Article: ] |
| 83. | Kudo T, Nurgalieva ZZ, Conner ME, Crawford S, Odenbreit S, Haas R, Graham DY, Yamaoka Y. Correlation between Helicobacter pylori OipA protein expression and oipA gene switch status. J Clin Microbiol. 2004;42:2279-2281. [PubMed] [Cited in This Article: ] |
| 84. | Lu H, Yamaoka Y, Graham DY. Helicobacter pylori virulence factors: facts and fantasies. Curr Opin Gastroenterol. 2005;21:653-659. [PubMed] [Cited in This Article: ] |
| 85. | Tabassam FH, Graham DY, Yamaoka Y. OipA plays a role in Helicobacter pylori-induced focal adhesion kinase activation and cytoskeletal re-organization. Cell Microbiol. 2008;10:1008-1020. [PubMed] [Cited in This Article: ] |
| 86. | Tabassam FH, Graham DY, Yamaoka Y. Helicobacter pylori activate epidermal growth factor receptor- and phosphatidylinositol 3-OH kinase-dependent Akt and glycogen synthase kinase 3beta phosphorylation. Cell Microbiol. 2009;11:70-82. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 82] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 87. | Tabassam FH, Graham DY, Yamaoka Y. Helicobacter pylori-associated regulation of forkhead transcription factors FoxO1/3a in human gastric cells. Helicobacter. 2012;17:193-202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 88. | Loh JT, Torres VJ, Algood HM, McClain MS, Cover TL. Helicobacter pylori HopQ outer membrane protein attenuates bacterial adherence to gastric epithelial cells. FEMS Microbiol Lett. 2008;289:53-58. [PubMed] [Cited in This Article: ] |
| 89. | Cao P, Cover TL. Two different families of hopQ alleles in Helicobacter pylori. J Clin Microbiol. 2002;40:4504-4511. [PubMed] [Cited in This Article: ] |
| 90. | Cao P, Lee KJ, Blaser MJ, Cover TL. Analysis of hopQ alleles in East Asian and Western strains of Helicobacter pylori. FEMS Microbiol Lett. 2005;251:37-43. [PubMed] [Cited in This Article: ] |
| 91. | Oleastro M, Santos A, Cordeiro R, Nunes B, Mégraud F, Ménard A. Clinical relevance and diversity of two homologous genes encoding glycosyltransferases in Helicobacter pylori. J Clin Microbiol. 2010;48:2885-2891. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 92. | Sicinschi LA, Correa P, Bravo LE, Peek RM, Wilson KT, Loh JT, Yepez MC, Gold BD, Thompson DT, Cover TL. Non-invasive genotyping of Helicobacter pylori cagA, vacA, and hopQ from asymptomatic children. Helicobacter. 2012;17:96-106. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 93. | Talebi Bezmin Abadi A, Mohabbati Mobarez A. High Prevalence of Helicobacter pylori hopQ II Genotype Isolated from Iranian Patients with Gastroduodenal Disorders. J Pathog. 2014;2014:842469. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 94. | Belogolova E, Bauer B, Pompaiah M, Asakura H, Brinkman V, Ertl C, Bartfeld S, Nechitaylo TY, Haas R, Machuy N. Helicobacter pylori outer membrane protein HopQ identified as a novel T4SS-associated virulence factor. Cell Microbiol. 2013;15:1896-1912. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 95. | Geis G, Leying H, Suerbaum S, Mai U, Opferkuch W. Ultrastructure and chemical analysis of Campylobacter pylori flagella. J Clin Microbiol. 1989;27:436-441. [PubMed] [Cited in This Article: ] |
| 96. | O’Toole PW, Lane MC, Porwollik S. Helicobacter pylori motility. Microbes Infect. 2000;2:1207-1214. [PubMed] [Cited in This Article: ] |
| 97. | Blair DF. Flagellar movement driven by proton translocation. FEBS Lett. 2003;545:86-95. [PubMed] [Cited in This Article: ] |
| 98. | Josenhans C, Labigne A, Suerbaum S. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J Bacteriol. 1995;177:3010-3020. [PubMed] [Cited in This Article: ] |
| 99. | Eaton KA, Suerbaum S, Josenhans C, Krakowka S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64:2445-2448. [PubMed] [Cited in This Article: ] |
| 100. | Kim JS, Chang JH, Chung SI, Yum JS. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J Bacteriol. 1999;181:6969-6976. [PubMed] [Cited in This Article: ] |
| 101. | Kavermann H, Burns BP, Angermuller K, Odenbreit S, Fischer W, Melchers K, Haas R. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J Exp Med. 2003;197:813-822. [PubMed] [Cited in This Article: ] |
| 102. | Asakura H, Churin Y, Bauer B, Boettcher JP, Bartfeld S, Hashii N, Kawasaki N, Mollenkopf HJ, Jungblut PR, Brinkmann V. Helicobacter pylori HP0518 affects flagellin glycosylation to alter bacterial motility. Mol Microbiol. 2010;78:1130-1144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 103. | Josenhans C, Vossebein L, Friedrich S, Suerbaum S. The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol Lett. 2002;210:165-172. [PubMed] [Cited in This Article: ] |
| 104. | Schirm M, Soo EC, Aubry AJ, Austin J, Thibault P, Logan SM. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol Microbiol. 2003;48:1579-1592. [PubMed] [Cited in This Article: ] |
| 105. | Forsyth MH, Atherton JC, Blaser MJ, Cover TL. Heterogeneity in levels of vacuolating cytotoxin gene (vacA) transcription among Helicobacter pylori strains. Infect Immun. 1998;66:3088-3094. [PubMed] [Cited in This Article: ] |
| 106. | van Doorn LJ, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, de Boer W, Quint W. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58-66. [PubMed] [Cited in This Article: ] |
| 107. | Peek RM, Thompson SA, Donahue JP, Tham KT, Atherton JC, Blaser MJ, Miller GG. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc Assoc Am Physicians. 1998;110:531-544. [PubMed] [Cited in This Article: ] |
| 108. | Shiota S, Suzuki R, Yamaoka Y. The significance of virulence factors in Helicobacter pylori. J Dig Dis. 2013;14:341-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 86] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 109. | Amjad N, Osman HA, Razak NA, Kassian J, Din J, bin Abdullah N. Clinical significance of Helicobacter pylori cagA and iceA genotype status. World J Gastroenterol. 2010;16:4443-4447. [PubMed] [Cited in This Article: ] |
| 110. | Boyanova L, Yordanov D, Gergova G, Markovska R, Mitov I. Association of iceA and babA genotypes in Helicobacter pylori strains with patient and strain characteristics. Antonie Van Leeuwenhoek. 2010;98:343-350. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 111. | Gatti LL, Módena JL, Payão SL, Smith Mde A, Fukuhara Y, Módena JL, de Oliveira RB, Brocchi M. Prevalence of Helicobacter pylori cagA, iceA and babA2 alleles in Brazilian patients with upper gastrointestinal diseases. Acta Trop. 2006;100:232-240. [PubMed] [Cited in This Article: ] |
| 112. | Tanih NF, McMillan M, Naidoo N, Ndip LM, Weaver LT, Ndip RN. Prevalence of Helicobacter pylori vacA, cagA and iceA genotypes in South African patients with upper gastrointestinal diseases. Acta Trop. 2010;116:68-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |