Published online Oct 14, 2014. doi: 10.3748/wjg.v20.i38.13648
Revised: February 4, 2014
Accepted: June 13, 2014
Published online: October 14, 2014
Several new treatment options for gastric cancer have been introduced but the prognosis of patients diagnosed with gastric cancer is still poor. Disease prognosis could be improved for high-risk individuals by implementing earlier screenings. Because many patients are asymptomatic during the early stages of gastric cancer, the diagnosis is often delayed and patients present with unresectable locally advanced or metastatic disease. Cytotoxic treatment has been shown to prolong survival in general, but not all patients are responders. The application of targeted therapies and multimodal treatment has improved prognosis for those with advanced disease. However, these new therapeutic strategies do not uniformly benefit all patients. Predicting whether patients will respond to specific therapies would be of particular value and would allow for stratifying patients for personalized treatment strategies. Metabolic imaging by positron emission tomography was the first technique with the potential to predict the response of esophago-gastric cancer to neoadjuvant therapy. Exploring and validating tissue-based biomarkers are ongoing processes. In this review, we discuss the status of several targeted therapies for gastric cancer, as well as proteomic and metabolic methods for investigating biomarkers for therapy response prediction in gastric cancer.
Core tip: The prognosis of patients diagnosed with gastric cancer is still poor. Cytotoxic treatment and targeted therapies have improved the prognosis of patients. However, patients do not benefit equally from these treatment options. The ability to predict whether patients will respond to specific therapies would be of particular value and would allow for stratifying patients for personalized treatment strategies. In this review, we discuss the status of targeted therapies for gastric cancer, as well as proteomic and metabolic methods for investigating biomarkers for therapy response prediction in gastric cancer.
- Citation: Aichler M, Luber B, Lordick F, Walch A. Proteomic and metabolic prediction of response to therapy in gastric cancer. World J Gastroenterol 2014; 20(38): 13648-13657
- URL: https://www.wjgnet.com/1007-9327/full/v20/i38/13648.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i38.13648
Gastric cancer morbidity ranks number four and mortality ranks number two with respect to worldwide cancer disease incidence and death[1]. Disease outcome depends on the tumor stage at the time of diagnosis; if not diagnosed early, prognosis is generally poor. Because most of the patients are asymptomatic during the early stages of gastric cancer, the diagnosis is often delayed and patients present with an unresectable locally advanced or metastatic disease. Current treatment protocols for gastric cancer are based primarily on results of clinical studies, and to a lesser degree on specific histological features. Treatment options for gastric cancer patients include surgery, chemotherapy and radiation therapy. The current prognosis for individuals with gastric cancer is grim, with fewer than 25% of patients surviving at 5 years after diagnosis[1,2]. Improved preoperative care and surgical techniques have produced clear benefits. However, real progress will only be achieved through the development of new treatment options that have reduced cell toxicity compared with that of standard therapeutic regimens. Recent studies are exploring approaches based on molecularly targeted therapeutics. This strategy personalizes the treatment therapy based on individual biomarkers, which can be used to select treatments that most effectively remediate cancers with specific biomarkers. Further work is needed to characterize early tumor responses to different neoadjuvant therapies.
Inter-individual variability of drug response or resistance but also individual tumor heterogeneity presents a challenge when treating gastric cancer. The identification of predictive tumor markers at the time of diagnosis that enable managers to develop more effective therapeutic strategies would be invaluable for patient treatment. Therefore, current research is focusing on indentifying novel, cancer- and patient-specific imaging and tissue- and blood-based biomarkers. Recent progress in gene sequencing and molecular diagnostics enables the identification of potentially useful biomarkers; however, many of these are controversial. Some studies are in apparent disagreement. Currently, the status of human epidermal growth factor receptor-2 (HER2) is used to select trastuzumab chemotherapy. However, no other biomarkers have been approved by medical consensus and governing agencies.
The quantification of molecular alterations correlating with heterogeneous gastric tumors at different stages of disease progression is technically challenging, and prevents the development of reliable biomarkers. Another obstacle is tumor heterogeneity, which is particularly evident in gastric cancer. New proteomic technologies are developing rapidly. Proteomic approaches promote large-scale sample screening and facilitate identification of proteins associated with disease and treatment. Metabolic changes associated with invasive cancers could be useful for predicting treatment responses; these changes could be tracked with specific metabolic tracers and molecular metabolic imaging during early assessment of patient-specific treatment strategies.
Gastric cancer is an active topic of clinical and basic research due to the high morbidity and mortality. A full understanding of molecular parameters that determine prognosis and how to predict and control therapeutic responses is lacking. Identification of specific biomarkers will elucidate the molecular, proteomic, and metabolic treatment responses and drug resistance mechanisms. This review discusses proteomic approaches for biomarker detection and metabolic imaging for early prediction of gastric cancer response to systemic and targeted therapies.
Due to recent large scale randomized studies, a globally accepted standard medical treatment of gastric cancer can be defined[3]. Recent studies have demonstrated that preoperative and perioperative chemotherapy improves the clinical outcome for patients with gastric cancer[4-6]. Patients with potentially resectable tumors are treated with surgery and perioperative chemotherapy or postoperative chemoradiation[7]. In the metastatic disease setting, patients are treated with combination chemotherapy because exposure to cytotoxins prolongs survival and improves control of symptoms[8]. Recently, an international author team evaluated commonly used therapeutic strategies for gastric cancer[3]. Their analysis indicated that a combination of cisplatin and 5-fluorouracil was the preferred strategy. However, oxaliplatin efficacy was equivalent to that of cisplatin[3], and oral fluoropyrimidines such as S-1 and capecitabine could be substituted for 5-fluorouracil[3]. Combination chemotherapy was preferred for the majority of patients due to the balanced benefit-to-risk ratio[3]. For fit patients with high tumor burden and possible secondary resectability, triplet chemotherapy had greater efficiency and produced higher treatment response rates[3]. Docetaxel resulted in significantly higher side effects[3]. For elderly and infirm patients, monotherapy and dose modifications were considered as beneficial[3]. In general, approximately 50% of patients respond to chemotherapy containing cisplatin, fluorouracil and anthracyclines or taxanes, but median survival is less than 12 mo with these combinations[9]. The optimal approach for a given patient remains unclear and controversial. However, one consistent finding is that patients who exhibit a histopathological response to neoadjuvant therapy are more likely to receive a survival benefit. Cytotoxic therapy provides positive response rates ranging from 20%-60%[10]. There are a few studies that evaluate clinical or histopathological markers that predict response and prognosis for neoadjuvant-treated gastric cancer, and none of the potential markers have been validated in prospective studies[10-15]. Identifying predictive, pretherapeutic markers for treatment response would facilitate customization of individual care strategies. Due to this patient specific situation, there is a need to use clinical, genomic, transcriptomic, proteomic, and other information sources to plot the optimal course for an individual patient in terms of disease risk assessment, prevention, treatment, or palliation. This is the concept of personalized medicine. Therapeutic approaches for gastric cancer will become increasingly customized in future clinical practice.
Research efforts for gastric cancer have focused on improving prognosis and decreasing chemotherapeutic toxicity. The addition of molecularly targeted agents to treatment protocols may achieve both of these goals. The human HER family is one of the main targets in human cancer therapies[16,17]. The HER family contains four related members, HER1 (ErbB1 or EGFR), HER2 (ErbB2), HER3 (ErbB3), and HER4 (ErbB4). HER2 is an important biomarker in gastric tumors and can be specifically targeted via a monoclonal antibody for trastuzumab therapy[18]. For patients with advanced gastric cancer or cancer of the gastro-esophageal junction, survival is improved with trastuzumab therapy combined with chemotherapy compared with that of chemotherapy alone[19]. Trastuzumab combined with capecitabine or 5-fluorouracil and cisplatin has been approved for treating patients with HER2-positive metastatic adenocarcinoma of the stomach or gastro-esophageal junction by the European Union, United States, and Japan.
Lapatinib, a tyrosine kinase inhibitor against both epidermal growth factor receptor (EGFR) and HER2, has had modest single activity[20]. Additionally, a statistically significant improvement in overall survival (primary endpoint) with the addition of lapatinib to capecitabine plus oxaliplatin (CapeOx) as the first-line treatment of advanced or metastatic gastric or gastro-esophageal adenocarcinoma could not be demonstrated (Logic trial)[21]. With regard to toxicity, lapatinib in combination with CapeOx showed an increased rate of grade 3 diarrhea and a higher rate of skin toxicity. In contrast to the success obtained with trastuzumab in advanced gastric cancer, monoclonal antibodies that target EGFR have failed to improve outcome in biologically unselected gastric cancer patients[22,23]. It remains to be elucidated from tumor tissue analyses if a small proportion of gastric cancer patients may benefit from anti-EGFR targeted therapy, e.g., in the case of EGFR gene amplification[24]. The negative results obtained with cetuximab (EXPAND study) and panitumumab (REAL3 study) emphasize the need to have a biologically meaningful target before studying targeted agents in larger populations. But the development of trastuzumab in HER2-overexpressing gastric cancer raises hope that further progress may be achieved.
Developing new targeted and multimodal therapies for gastrointestinal cancer has improved patient prognosis. However, additional treatment choices add greater complexity to the therapeutic strategy, and selecting the correct regimen has become more challenging for clinical managers. Therefore, the identification of new therapeutic response biomarkers is crucial.
The selection of anticancer regimens based on individual patient biomarkers constitutes personalized cancer treatment. There is a strong need to identify parameters that can be used as reporters of tumor responsiveness during the early phases of neoadjuvant therapies. Current therapeutic management is based primarily on clinical data and histological features. Several new treatment options have been introduced recently, but variations in individual responses and drug resistance present challenges. Many promising markers for disease prognosis or therapeutic response have been identified, but the diagnostic value of many biomarkers is controversial. The only molecular marker currently in clinical use to tailor patient therapy is the HER2 status for treatment with trastuzumab. Identifying new cancer-specific biomarkers for predicting patient responses to different therapies is currently a focus of translational research. Table 1 presents studies that have identified potential biomarkers that could be used for predicting therapy response in gastric cancer patients[25-33]. These “discovery” studies provide some perspectives for establishment of further biomarkers. In this regard, in a recently published study it was demonstrated that high levels of serum AMBP (Alpha-1 Microglobulin/Bikunin Precursor) could predict the poor response of gastric cancer patients treated with paclitaxel-capecitabine chemotherapy[25]. Also recently published was a study identifying REG Iα (Regenerating Gene Iα) expression in tissue biopsies of gastric cancer patients for predicting response to chemotherapy with S1 plus cisplatin[26]. Another potential biomarker, FoxM1 (Forkhead Box M1 Transcription Factor), was suggested as biomarker for resistance to chemotherapy with docetaxel in addition to 5-fluorouracil derivate plus cisplatin when overexpressed gastric cancer tissue[27]. The methylation of the apoptosis-related genes, BNIP3 (Bcl-2/Adenovirus E1B 19 kDa-interacting Protein 3) and DAPK (Death Associated Protein Kinase) was examined in tumor samples from patients treated with fluoropyrimidine-based chemotherapy for metastatic or recurrent gastric cancer and was found to indicate lower response to the chemotherapy[28]. High levels of REG IV (regenerating gene family, member 4) in the serum of gastric cancer patients were identified to predict resistance to 5-fluorouracil-based chemotherapy[29].
| Ref. | Biomarker | Sample | Patients | Chemotherapy |
| Huang et al[25], 2013 | AMBP | Serum | 17 GC patients | paclitaxel, capecitabine |
| Sekikawa et al[26], 2013 | REG Iα | Tissue | 70 GC patients | S-1, Cisplatin |
| Okada et al[27], 2013 | FOXM1 | Tissue | 81 GC patients | docetaxel, 5-FU, cisplatin/5-FU, cisplatin |
| Sugita et al[28], 2010 | BNIP3 | Tissue | 80 GC patients | 5-FU, irinotecan/docetaxel/cisplatin |
| DAPK | ||||
| Mitani et al[29], 2007 | REG4 | Serum | 36 GC patients | 5-FU, cisplatin |
These new biomarkers have not yet progressed from basic research into clinical practice. A greater understanding of these biomarkers could reveal novel insights into the molecular changes underlying cancer progression, metabolic responses to treatments, and mechanisms leading to chemotherapy resistance. Metabolic and proteomic changes are features of invasive cancers, and may provide valuable information for assessing prognosis and response to treatment for patients with gastric cancer. Proteomics evaluates protein expression, post-translational modifications, and complex expression patterns in tissues, cells, and biological fluids[34,35].
Changes in protein profiles reflect changes in cellular metabolism and cellular responses to extracellular conditions. Proteins are key effector molecules that influence pathological conditions. The development of proteomics technologies enables screening of different samples such as fluids and clinical tissues, including fresh/frozen and formalin-fixed paraffin embedded (FFPE) materials. Fresh/frozen tissue is more suitable for proteomics analysis than chemically cross-linked material. However, archival FFPE clinical samples represent a rich source for proteomics investigations, and these are often linked with extensive follow-up patient information that report disease outcomes. Independent studies demonstrate that frozen samples are equivalent to chemically fixed samples after rehydration and heat-induced reversal of fixation[36-38]. Obvious advantages of FFPE samples are convenience of handling, storage, and archival follow-up information that often covers decades. Therefore, FFPE material has been used for cancer research by many groups[38-44].
Proteomics studies can provide information about general protein expression patterns, expression of individual proteins, post-translational protein modification, and protein-protein interactions. Proteins can be analyzed by electrophoresis, chromatography, visualization, and mass spectrometry. The development of advanced protein separation systems such as high-resolution chromatography and high-sensitivity mass spectrometry have facilitated development of proteomics technologies[45]. Proteomics studies using mass spectrometry techniques are discovering important data that can be used to predict therapy responses, particularly studies using specialized protein separation techniques, matrix-assisted laser desorption-ionization (MALDI), and time-of-flight mass spectrometry (TOF-MS)[46,47]. Mass spectrometry-based proteomics is generally performed on fresh/frozen tissues. Mass spectrometry of FFPE tissues requires proteolytic digestion of the samples to generate peptides that can be analyzed by liquid chromatography−mass spectrometry (LC-MS). Applications of LC-MS for analysis of FFPE samples have recently been reviewed by Steiner et al[48]. These identified protein profiles can be used for analysis of useful biomarkers for gastric cancer. A recently published workflow for analysis of FFPE samples of colon adenomas demonstrated that it is possible to analyze the proteome from microdissected tissue samples[49]. Recent progress in mass spectrometry techniques may lead to quantitative, reproducible, and highly multiplex proteomics analyses of FFPE samples in the future.
Mass spectrometry detects and identifies the chemical composition of samples on the basis of their mass-to-charge ratio (m/z) after ionization, and can be used to determine protein molecular weight, structure, and posttranslational modifications. In TOF-MS, ionic flight times are measured over a fixed distance and correlated with specific m/z values. The measured output counts the total number of ions at each m/z value. MALDI-TOF analysis is highly sensitive and accurate, even for proteins with molecular weights less than 200 kDa[50]. Coupling mass spectrometry with protein separation methods enables characterization of amino acid sequences and post-translational modification. Mass spectrometry is a powerful tool for proteomics analysis, and has been used to identify biomarkers in cancer proteomes for early diagnosis, to assess disease prognosis, and to predict therapeutic responses[51-53]. Several biomarker studies have been conducted using serum samples. The main obstacle for proteomic analysis of serum is the large variability in protein concentrations that can render identification of the low-abundance proteins of interest extremely challenging. Serum screening may identify non-specific markers associated with systemic responses or secondary processes unrelated to cancer. These can include effects from diet, smoking, alcohol consumption, or other factors that complicate analysis. These serum-related issues are not encountered when performing mass spectrometry analysis of tissue samples.
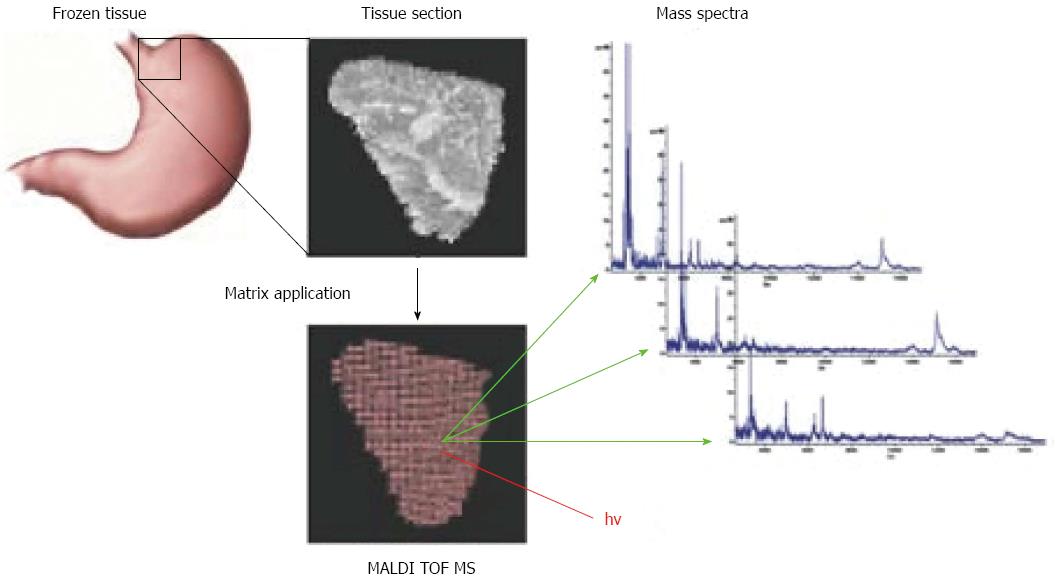
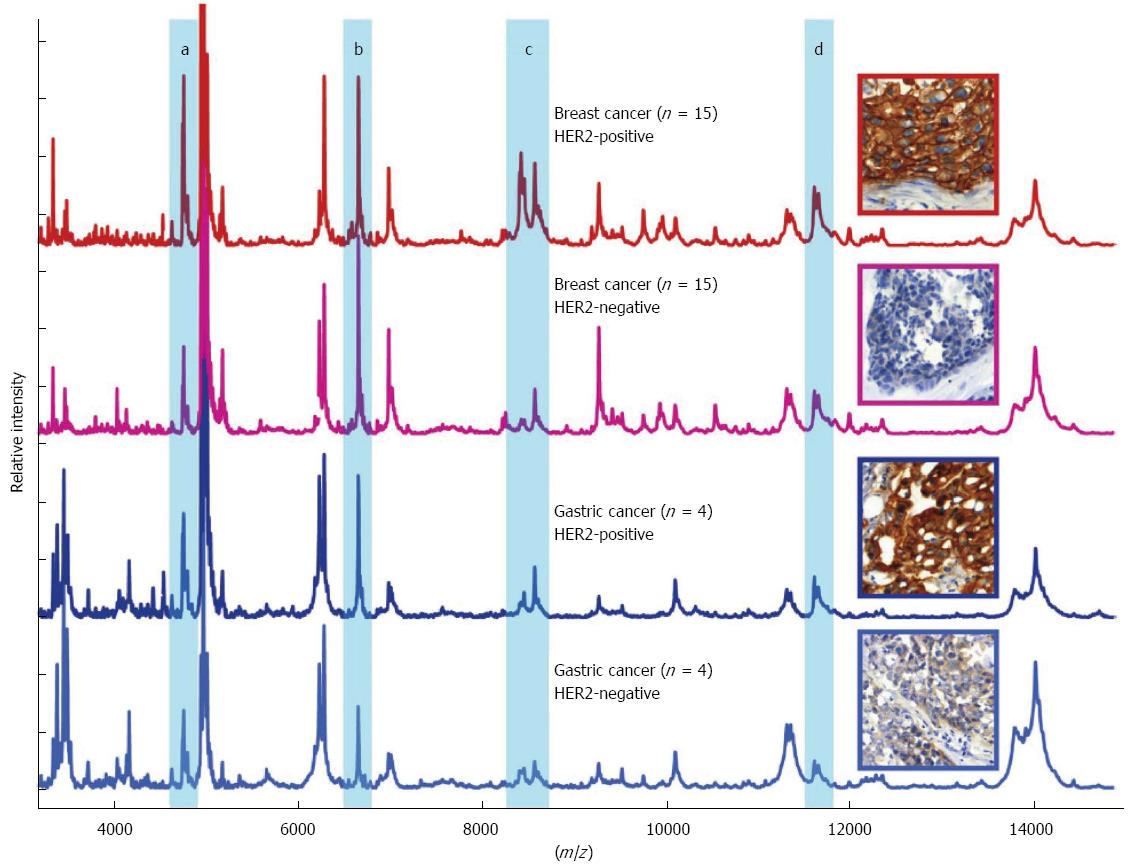
A novel method has recently been developed for cancer-specific biomarker screening of patient tissue specimens. The MALDI imaging mass spectrometry technology facilitates the application of MALDI mass spectrometry to the analysis of tissue sections (Figure 1)[50,54-59]. In MALDI imaging mass spectrometry, the molecular content identified by mass spectrometry is specifically localized within tissue sections and biopsies. Peptides, proteins, posttranslational modifications, therapeutic agents and their metabolites, lipids, cell metabolites, molecular tracers, contrast agents, and toxins can be identified and localized[58]. This technique correlates in situ molecular patterns with m/z distributions. Tissue sections are scanned and a mass spectrum is acquired for selected regions, which are then subjected to histomorphological analysis and visualization (Figure 1). MALDI imaging has been implemented in several studies to identify differential protein expression profiles for human glioblastoma cancer, non-small-cell lung tumors, and ovarian cancer[54,60,61]. One of the first MALDI Imaging studies in gastric cancer demonstrated that MALDI imaging in combination with hierarchical clustering of the m/z values allows the comprehensive analysis of the in situ cancer proteome[62]. This cluster analysis allowed for the classification of complex human tissues and facilitated specific and cancer-related in situ biomarker analysis and identification. Also, in the case of gastric cancer, MALDI Imaging could be obtained as a diagnostic tool to identify early-stage tumor. By histology-directed profiling of 63 gastric cancer and 43 healthy endoscopic biopsies, profiles for separating tumor from healthy tissue, and for distinguishing stage Ia from more advanced stages were identified[63]. The results from this study could be of clinical relevance, because stage Ia lesions are potential candidates for endoscopic treatment. For patients with more advanced-stage disease, clinically relevant information is related to improving risk stratification. A recent study analyzed 63 intestinal primary gastric cancer tissues using MALDI imaging[64] and identified 7 tumor-specific proteins that independently correlated with poor survival. A previously unknown protein, CRIP1, was identified and confirmed to be an independent prognostic factor for gastric cancer. A proof-of-principle MALDI imaging study demonstrated that the HER2 status of gastric cancer could be predicted accurately by specific protein patterns (Figure 2)[65]. A recently published MALDI Imaging study demonstrated that the clinical response to neoadjuvant chemotherapy with cisplatin and 5-FU in adenocarcinomas of the gastro-esophageal junction could be correlated to preexisting defects in mitochondria of the patients’ tumor cells[66]. Additionally in this study several mitochondrial proteins were identified which previously have not been recognized in the context of neoadjuvant chemotherapy treatment. In general, because of its practical simplicity and ability to gain reliable information, even from endoscopic biopsy sections, MALDI Imaging might have the potential to complement histopathological evaluation for assisting diagnostic, risk assessment, or response prediction to therapy.
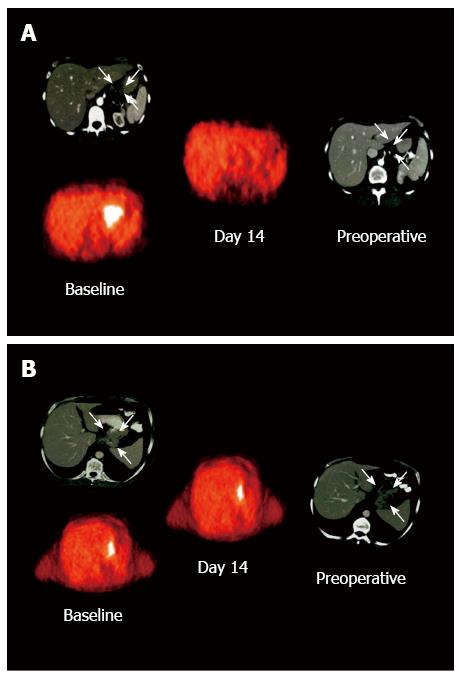
Molecular imaging of tumor metabolism using specific tracers could guide decisions about treatment strategies for cancer patients. Since the discovery regarding glucose metabolism in cancerous tumors by Warburg, there is consensus that malignant cell metabolism is crucial for pathogenesis and progression of cancer[67]. Changes in glucose metabolism are determined using fluorodeoxyglucose (FDG)-positron emission tomography (PET) and positron emission tomography-computed tomography (PET-CT) imaging. These changes can be used for tumor diagnosis. Measurement of tumor FDG uptake using PET can facilitate the assessment of tumor cell metabolic activity in vivo. 18F-FDG-PET can be used to measure the response to therapy, tumor metabolism, and patient prognosis. PET imaging can determine the tumor response to treatment during the course of chemotherapy, radiotherapy, or chemoradiotherapy. Changes in 18F-FDG uptake compared with that in pretherapeutic scans can be correlated with histopathological changes and/or survival. During the early course of treatment, tumor metabolic activity is significantly reduced in patients that positively respond to treatment compared with those that do not (Figure 3)[68,69].
Early assessment of treatment response for gastric cancer using FDG-PET is challenging because many primary tumors are not FDG avid[70-74]. If the tumor is FDG avid, prediction of response and prognosis using FDG-PET is feasible for gastric cancer[75]. Ott et al[75] established a standard for assessing if a treatment resulted in positive patient responses and improved patient prognoses; after two weeks of chemotherapy, approximately 35% of patients should have reduced FDG uptake compared with that of the pre-treatment FDG uptake. This standard was corroborated by a subsequent study with a larger patient cohort, in which approximately 33% of patients had insufficient FDG to monitor using FDG-PET[76]. Survival data identified three independent prognostic groups, including metabolic responders, metabolic non-responders, and non FDG-avid patients. Herrmann et al[77] investigated whether a marker of tumor cell proliferation, 18F-fluorothymidine (FLT), could be used to detect locally advanced gastric cancer. Absolute uptake values for 18F-FLT were lower than those for 18F-FDG, but 18F-FLT-PET exhibited higher sensitivity. Therefore, 18F-FLT-PET may be a useful diagnostic tool for quantifying tumor cell proliferation. A recently published prospective study by Ott et al[78] reported that non-FDG-avid gastric tumors can be visualized with the proliferation marker FLT. This can expand the potential of molecular imaging for assessing responses to neoadjuvant therapy.
Gastric cancer is a biological heterogeneous disease; therefore, no single medical treatment is the best option for all types of gastric cancer. Even for the treatment with classical cytotoxic therapies, different sensitivities to specific agents probably exist in different gastric cancer subtypes. Current treatment protocols for gastric cancer are based primarily on clinical data and histological features. Therefore, new targeted agents are needed beside the already established anti-HER2 directed treatment with trastuzumab. Several potential biomarkers have been identified that can predict treatment responses, and can be used to customize therapeutic approaches with respect to specific tumor parameters.
With a better proteomic and metabolic characterization of gastric cancer, new and improved treatment options may become available in future. To identify patients that could benefit most from novel treatments, it is important to assess early patient responses. Molecular and proteomics analyses have identified a number of proteins that might be useful for predicting therapeutic responses. Most of these biomarkers require further validation in larger studies. Molecular imaging could facilitate early assessment of patient responses to treatments. MALDI imaging is a novel approach to identify new biomarkers. The combination of different approaches is necessary to identify new, cancer-specific, and patient-specific biomarkers that could be used to establish personalized treatment strategies and clinical management of patients with gastric cancer.
P- Reviewer: Ierardi E S- Editor: Ma YJ L- Editor: A E- Editor: Ma S
| 1. | Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9215] [Cited by in F6Publishing: 9719] [Article Influence: 883.5] [Reference Citation Analysis (3)] |
| 2. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11128] [Cited by in F6Publishing: 11614] [Article Influence: 893.4] [Reference Citation Analysis (4)] |
| 3. | Lordick F, Lorenzen S, Yamada Y, Ilson D. Optimal chemotherapy for advanced gastric cancer: is there a global consensus? Gastric Cancer. 2014;17:213-225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 4. | Ott K, Sendler A, Becker K, Dittler HJ, Helmberger H, Busch R, Kollmannsberger C, Siewert JR, Fink U. Neoadjuvant chemotherapy with cisplatin, 5-FU, and leucovorin (PLF) in locally advanced gastric cancer: a prospective phase II study. Gastric Cancer. 2003;6:159-167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 107] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 5. | Schuhmacher C, Gretschel S, Lordick F, Reichardt P, Hohenberger W, Eisenberger CF, Haag C, Mauer ME, Hasan B, Welch J. Neoadjuvant chemotherapy compared with surgery alone for locally advanced cancer of the stomach and cardia: European Organisation for Research and Treatment of Cancer randomized trial 40954. J Clin Oncol. 2010;28:5210-5218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 511] [Cited by in F6Publishing: 496] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 6. | Sakuramoto S, Sasako M, Yamaguchi T, Kinoshita T, Fujii M, Nashimoto A, Furukawa H, Nakajima T, Ohashi Y, Imamura H. Adjuvant chemotherapy for gastric cancer with S-1, an oral fluoropyrimidine. N Engl J Med. 2007;357:1810-1820. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1771] [Cited by in F6Publishing: 1821] [Article Influence: 107.1] [Reference Citation Analysis (0)] |
| 7. | Macdonald JS, Smalley SR, Benedetti J, Hundahl SA, Estes NC, Stemmermann GN, Haller DG, Ajani JA, Gunderson LL, Jessup JM. Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. N Engl J Med. 2001;345:725-730. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2465] [Cited by in F6Publishing: 2365] [Article Influence: 102.8] [Reference Citation Analysis (0)] |
| 8. | Van Cutsem E, Moiseyenko VM, Tjulandin S, Majlis A, Constenla M, Boni C, Rodrigues A, Fodor M, Chao Y, Voznyi E. Phase III study of docetaxel and cisplatin plus fluorouracil compared with cisplatin and fluorouracil as first-line therapy for advanced gastric cancer: a report of the V325 Study Group. J Clin Oncol. 2006;24:4991-4997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1331] [Cited by in F6Publishing: 1405] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 9. | Jiang Y, Ajani JA. Multidisciplinary management of gastric cancer. Curr Opin Gastroenterol. 2010;26:640-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 36] [Article Influence: 2.6] [Reference Citation Analysis (1)] |
| 10. | Persiani R, D’Ugo D, Rausei S, Sermoneta D, Barone C, Pozzo C, Ricci R, La Torre G, Picciocchi A. Prognostic indicators in locally advanced gastric cancer (LAGC) treated with preoperative chemotherapy and D2-gastrectomy. J Surg Oncol. 2005;89:227-36; discussion 237-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 30] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 11. | Rohatgi PR, Mansfield PF, Crane CH, Wu TT, Sunder PK, Ross WA, Morris JS, Pisters PW, Feig BW, Gunderson LL. Surgical pathology stage by American Joint Commission on Cancer criteria predicts patient survival after preoperative chemoradiation for localized gastric carcinoma. Cancer. 2006;107:1475-1482. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 12. | Ninomiya Y, Yanagisawa A, Kato Y, Kitagawa T, Ishihara S, Nakajima T. Histological indications of a favorable prognosis with far-advanced gastric carcinomas after preoperative chemotherapy. J Cancer Res Clin Oncol. 1999;125:699-706. [PubMed] [Cited in This Article: ] |
| 13. | Nagashima F, Boku N, Ohtsu A, Yoshida S, Hasebe T, Ochiai A, Sakata Y, Saito H, Miyata Y, Hyodo I. Biological markers as a predictor for response and prognosis of unresectable gastric cancer patients treated with irinotecan and cisplatin. Jpn J Clin Oncol. 2005;35:714-719. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 14. | Jhawer M, Coit D, Brennan M, Qin LX, Gonen M, Klimstra D, Tang L, Kelsen DP, Shah MA. Perineural invasion after preoperative chemotherapy predicts poor survival in patients with locally advanced gastric cancer: gene expression analysis with pathologic validation. Am J Clin Oncol. 2009;32:356-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Mansour JC, Tang L, Shah M, Bentrem D, Klimstra DS, Gonen M, Kelsen DP, Brennan MF, Coit DG. Does graded histologic response after neoadjuvant chemotherapy predict survival for completely resected gastric cancer? Ann Surg Oncol. 2007;14:3412-3418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 89] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 16. | Fornaro L, Lucchesi M, Caparello C, Vasile E, Caponi S, Ginocchi L, Masi G, Falcone A. Anti-HER agents in gastric cancer: from bench to bedside. Nat Rev Gastroenterol Hepatol. 2011;8:369-383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | de Mello RA, Marques AM, Araújo A. HER2 therapies and gastric cancer: a step forward. World J Gastroenterol. 2013;19:6165-6169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 34] [Cited by in F6Publishing: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 18. | Gravalos C, Jimeno A. HER2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann Oncol. 2008;19:1523-1529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 737] [Cited by in F6Publishing: 809] [Article Influence: 50.6] [Reference Citation Analysis (2)] |
| 19. | Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687-697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4615] [Cited by in F6Publishing: 4831] [Article Influence: 345.1] [Reference Citation Analysis (1)] |
| 20. | Iqbal S, Goldman B, Fenoglio-Preiser CM, Lenz HJ, Zhang W, Danenberg KD, Shibata SI, Blanke CD. Southwest Oncology Group study S0413: a phase II trial of lapatinib (GW572016) as first-line therapy in patients with advanced or metastatic gastric cancer. Ann Oncol. 2011;22:2610-2615. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 109] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 21. | Hecht JR, Qin S. Lapatinib in combination with capecitabine plus oxaliplatin (CapeOx) in HER2-positive advanced or metastatic gastric, esophageal, or gastroesophageal adenocarcinoma (AC): The TRIO-031/LOGIC Trial. J Clin Oncol. 2013;13 suppl:abstr LBA4001. [Cited in This Article: ] |
| 22. | Lordick F, Kang YK, Chung HC, Salman P, Oh SC, Bodoky G, Kurteva G, Volovat C, Moiseyenko VM, Gorbunova V. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): a randomised, open-label phase 3 trial. Lancet Oncol. 2013;14:490-499. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 589] [Cited by in F6Publishing: 640] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 23. | Waddell T, Chau I, Cunningham D, Gonzalez D, Okines AF, Okines C, Wotherspoon A, Saffery C, Middleton G, Wadsley J. Epirubicin, oxaliplatin, and capecitabine with or without panitumumab for patients with previously untreated advanced oesophagogastric cancer (REAL3): a randomised, open-label phase 3 trial. Lancet Oncol. 2013;14:481-489. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 504] [Cited by in F6Publishing: 554] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 24. | Luber B, Deplazes J, Keller G, Walch A, Rauser S, Eichmann M, Langer R, Höfler H, Hegewisch-Becker S, Folprecht G. Biomarker analysis of cetuximab plus oxaliplatin/leucovorin/5-fluorouracil in first-line metastatic gastric and oesophago-gastric junction cancer: results from a phase II trial of the Arbeitsgemeinschaft Internistische Onkologie (AIO). BMC Cancer. 2011;11:509. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 25. | Huang H, Han Y, Gao J, Feng J, Zhu L, Qu L, Shen L, Shou C. High level of serum AMBP is associated with poor response to paclitaxel-capecitabine chemotherapy in advanced gastric cancer patients. Med Oncol. 2013;30:748. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 26. | Sekikawa A, Fukui H, Zhang X, Maruo T, Tsumura T, Okabe Y, Wakasa T, Osaki Y, Chiba T, Tomita T. REG Iα is a biomarker for predicting response to chemotherapy with S-1 plus cisplatin in patients with unresectable stage IV gastric cancer. Br J Cancer. 2013;108:395-401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | Okada K, Fujiwara Y, Takahashi T, Nakamura Y, Takiguchi S, Nakajima K, Miyata H, Yamasaki M, Kurokawa Y, Mori M. Overexpression of forkhead box M1 transcription factor (FOXM1) is a potential prognostic marker and enhances chemoresistance for docetaxel in gastric cancer. Ann Surg Oncol. 2013;20:1035-1043. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 28. | Sugita H, Iida S, Inokuchi M, Kato K, Ishiguro M, Ishikawa T, Takagi Y, Enjoji M, Yamada H, Uetake H. Methylation of BNIP3 and DAPK indicates lower response to chemotherapy and poor prognosis in gastric cancer. Oncol Rep. 2011;25:513-518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 29. | Mitani Y, Oue N, Matsumura S, Yoshida K, Noguchi T, Ito M, Tanaka S, Kuniyasu H, Kamata N, Yasui W. Reg IV is a serum biomarker for gastric cancer patients and predicts response to 5-fluorouracil-based chemotherapy. Oncogene. 2007;26:4383-4393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 101] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 30. | Su Y, Shen J, Qian H, Ma H, Ji J, Ma H, Ma L, Zhang W, Meng L, Li Z. Diagnosis of gastric cancer using decision tree classification of mass spectral data. Cancer Sci. 2007;98:37-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 62] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 31. | Qian HG, Shen J, Ma H, Ma HC, Su YH, Hao CY, Xing BC, Huang XF, Shou CC. Preliminary study on proteomics of gastric carcinoma and its clinical significance. World J Gastroenterol. 2005;11:6249-6253. [PubMed] [Cited in This Article: ] |
| 32. | Lim JY, Cho JY, Paik YH, Chang YS, Kim HG. Diagnostic application of serum proteomic patterns in gastric cancer patients by ProteinChip surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Int J Biol Markers. 2007;22:281-286. [PubMed] [Cited in This Article: ] |
| 33. | Poon TC, Sung JJ, Chow SM, Ng EK, Yu AC, Chu ES, Hui AM, Leung WK. Diagnosis of gastric cancer by serum proteomic fingerprinting. Gastroenterology. 2006;130:1858-1864. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 48] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 34. | Veenstra TD, Conrads TP, Hood BL, Avellino AM, Ellenbogen RG, Morrison RS. Biomarkers: mining the biofluid proteome. Mol Cell Proteomics. 2005;4:409-418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 192] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 35. | Röcken C, Ketterlinus R, Ebert MP. Application of proteome analysis to the assessment of prognosis and response prediction in clinical oncology. Curr Cancer Drug Targets. 2008;8:141-145. [PubMed] [Cited in This Article: ] |
| 36. | Bateman NW, Sun M, Bhargava R, Hood BL, Darfler MM, Kovatich AJ, Hooke JA, Krizman DB, Conrads TP. Differential proteomic analysis of late-stage and recurrent breast cancer from formalin-fixed paraffin-embedded tissues. J Proteome Res. 2011;10:1323-1332. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 37. | Ostasiewicz P, Zielinska DF, Mann M, Wiśniewski JR. Proteome, phosphoproteome, and N-glycoproteome are quantitatively preserved in formalin-fixed paraffin-embedded tissue and analyzable by high-resolution mass spectrometry. J Proteome Res. 2010;9:3688-3700. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 184] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 38. | Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, Ringeisen BR, Sesterhenn IA, Conrads TP, Veenstra TD, Krizman DB. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol Cell Proteomics. 2005;4:1741-1753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 220] [Cited by in F6Publishing: 201] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 39. | Shi SR, Liu C, Balgley BM, Lee C, Taylor CR. Protein extraction from formalin-fixed, paraffin-embedded tissue sections: quality evaluation by mass spectrometry. J Histochem Cytochem. 2006;54:739-743. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 173] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 40. | Jiang X, Jiang X, Feng S, Tian R, Ye M, Zou H. Development of efficient protein extraction methods for shotgun proteome analysis of formalin-fixed tissues. J Proteome Res. 2007;6:1038-1047. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 79] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 41. | Guo T, Wang W, Rudnick PA, Song T, Li J, Zhuang Z, Weil RJ, DeVoe DL, Lee CS, Balgley BM. Proteome analysis of microdissected formalin-fixed and paraffin-embedded tissue specimens. J Histochem Cytochem. 2007;55:763-772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 98] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 42. | Nirmalan NJ, Hughes C, Peng J, McKenna T, Langridge J, Cairns DA, Harnden P, Selby PJ, Banks RE. Initial development and validation of a novel extraction method for quantitative mining of the formalin-fixed, paraffin-embedded tissue proteome for biomarker investigations. J Proteome Res. 2011;10:896-906. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 43. | Wolff C, Schott C, Porschewski P, Reischauer B, Becker KF. Successful protein extraction from over-fixed and long-term stored formalin-fixed tissues. PLoS One. 2011;6:e16353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 44. | Magdeldin S, Yamamoto T. Toward deciphering proteomes of formalin-fixed paraffin-embedded (FFPE) tissues. Proteomics. 2012;12:1045-1058. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 45. | Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395-398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 330] [Cited by in F6Publishing: 296] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 46. | Herosimczyk A, Dejeans N, Sayd T, Ozgo M, Skrzypczak WF, Mazur A. Plasma proteome analysis: 2D gels and chips. J Physiol Pharmacol. 2006;57 Suppl 7:81-93. [PubMed] [Cited in This Article: ] |
| 47. | López JL. Two-dimensional electrophoresis in proteome expression analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;849:190-202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 98] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 48. | Steiner C, Ducret A, Tille JC, Thomas M, McKee TA, Rubbia-Brandt L, Scherl A, Lescuyer P, Cutler P. Applications of mass spectrometry for quantitative protein analysis in formalin-fixed paraffin-embedded tissues. Proteomics. 2014;14:441-451. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 49. | Wiśniewski JR, Duś K, Mann M. Proteomic workflow for analysis of archival formalin-fixed and paraffin-embedded clinical samples to a depth of 10 000 proteins. Proteomics Clin Appl. 2013;7:225-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 120] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 50. | Chaurand P, Schwartz SA, Billheimer D, Xu BJ, Crecelius A, Caprioli RM. Integrating histology and imaging mass spectrometry. Anal Chem. 2004;76:1145-1155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 288] [Cited by in F6Publishing: 258] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 51. | Leman ES, Schoen RE, Magheli A, Sokoll LJ, Chan DW, Getzenberg RH. Evaluation of colon cancer-specific antigen 2 as a potential serum marker for colorectal cancer. Clin Cancer Res. 2008;14:1349-1354. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 52. | Ward DG, Suggett N, Cheng Y, Wei W, Johnson H, Billingham LJ, Ismail T, Wakelam MJ, Johnson PJ, Martin A. Identification of serum biomarkers for colon cancer by proteomic analysis. Br J Cancer. 2006;94:1898-1905. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 161] [Article Influence: 8.9] [Reference Citation Analysis (1)] |
| 53. | Xia Q, Kong XT, Zhang GA, Hou XJ, Qiang H, Zhong RQ. Proteomics-based identification of DEAD-box protein 48 as a novel autoantigen, a prospective serum marker for pancreatic cancer. Biochem Biophys Res Commun. 2005;330:526-532. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 50] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 54. | Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med. 2001;7:493-496. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 966] [Cited by in F6Publishing: 824] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 55. | Walch A, Rauser S, Deininger SO, Höfler H. MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology. Histochem Cell Biol. 2008;130:421-434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 260] [Cited by in F6Publishing: 200] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 56. | Fournier I, Wisztorski M, Salzet M. Tissue imaging using MALDI-MS: a new frontier of histopathology proteomics. Expert Rev Proteomics. 2008;5:413-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 63] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 57. | Wisztorski M, Croix D, Macagno E, Fournier I, Salzet M. Molecular MALDI imaging: an emerging technology for neuroscience studies. Dev Neurobiol. 2008;68:845-858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 58. | Balluff B, Rauser S, Ebert MP, Siveke JT, Höfler H, Walch A. Direct molecular tissue analysis by MALDI imaging mass spectrometry in the field of gastrointestinal disease. Gastroenterology. 2012;143:544-9.e1-544-9.e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 59. | Norris JL, Caprioli RM. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev. 2013;113:2309-2342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 557] [Cited by in F6Publishing: 488] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 60. | Yanagisawa K, Shyr Y, Xu BJ, Massion PP, Larsen PH, White BC, Roberts JR, Edgerton M, Gonzalez A, Nadaf S. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362:433-439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 487] [Cited by in F6Publishing: 503] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 61. | Lemaire R, Menguellet SA, Stauber J, Marchaudon V, Lucot JP, Collinet P, Farine MO, Vinatier D, Day R, Ducoroy P. Specific MALDI imaging and profiling for biomarker hunting and validation: fragment of the 11S proteasome activator complex, Reg alpha fragment, is a new potential ovary cancer biomarker. J Proteome Res. 2007;6:4127-4134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 171] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 62. | Deininger SO, Ebert MP, Fütterer A, Gerhard M, Röcken C. MALDI imaging combined with hierarchical clustering as a new tool for the interpretation of complex human cancers. J Proteome Res. 2008;7:5230-5236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 198] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 63. | Kim HK, Reyzer ML, Choi IJ, Kim CG, Kim HS, Oshima A, Chertov O, Colantonio S, Fisher RJ, Allen JL. Gastric cancer-specific protein profile identified using endoscopic biopsy samples via MALDI mass spectrometry. J Proteome Res. 2010;9:4123-4130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 64. | Balluff B, Rauser S, Meding S, Elsner M, Schöne C, Feuchtinger A, Schuhmacher C, Novotny A, Jütting U, Maccarrone G. MALDI imaging identifies prognostic seven-protein signature of novel tissue markers in intestinal-type gastric cancer. Am J Pathol. 2011;179:2720-2729. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 116] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 65. | Balluff B, Elsner M, Kowarsch A, Rauser S, Meding S, Schuhmacher C, Feith M, Herrmann K, Röcken C, Schmid RM. Classification of HER2/neu status in gastric cancer using a breast-cancer derived proteome classifier. J Proteome Res. 2010;9:6317-6322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 66. | Aichler M, Elsner M, Ludyga N, Feuchtinger A, Zangen V, Maier SK, Balluff B, Schöne C, Hierber L, Braselmann H. Clinical response to chemotherapy in oesophageal adenocarcinoma patients is linked to defects in mitochondria. J Pathol. 2013;230:410-419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 67. | Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269-270. [PubMed] [Cited in This Article: ] |
| 68. | Weber WA, Wieder H. Monitoring chemotherapy and radiotherapy of solid tumors. Eur J Nucl Med Mol Imaging. 2006;33 Suppl 1:27-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 97] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 69. | Weber WA, Figlin R. Monitoring cancer treatment with PET/CT: does it make a difference? J Nucl Med. 2007;48 Suppl 1:36S-44S. [PubMed] [Cited in This Article: ] |
| 70. | Chen J, Cheong JH, Yun MJ, Kim J, Lim JS, Hyung WJ, Noh SH. Improvement in preoperative staging of gastric adenocarcinoma with positron emission tomography. Cancer. 2005;103:2383-2390. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 168] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 71. | Kim SK, Kang KW, Lee JS, Kim HK, Chang HJ, Choi JY, Lee JH, Ryu KW, Kim YW, Bae JM. Assessment of lymph node metastases using 18F-FDG PET in patients with advanced gastric cancer. Eur J Nucl Med Mol Imaging. 2006;33:148-155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 126] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 72. | Mochiki E, Kuwano H, Katoh H, Asao T, Oriuchi N, Endo K. Evaluation of 18F-2-deoxy-2-fluoro-D-glucose positron emission tomography for gastric cancer. World J Surg. 2004;28:247-253. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 161] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 73. | Yoshioka T, Yamaguchi K, Kubota K, Saginoya T, Yamazaki T, Ido T, Yamaura G, Takahashi H, Fukuda H, Kanamaru R. Evaluation of 18F-FDG PET in patients with advanced, metastatic, or recurrent gastric cancer. J Nucl Med. 2003;44:690-699. [PubMed] [Cited in This Article: ] |
| 74. | Yun M, Lim JS, Noh SH, Hyung WJ, Cheong JH, Bong JK, Cho A, Lee JD. Lymph node staging of gastric cancer using (18)F-FDG PET: a comparison study with CT. J Nucl Med. 2005;46:1582-1588. [PubMed] [Cited in This Article: ] |
| 75. | Ott K, Fink U, Becker K, Stahl A, Dittler HJ, Busch R, Stein H, Lordick F, Link T, Schwaiger M. Prediction of response to preoperative chemotherapy in gastric carcinoma by metabolic imaging: results of a prospective trial. J Clin Oncol. 2003;21:4604-4610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 256] [Cited by in F6Publishing: 268] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 76. | Ott K, Herrmann K, Lordick F, Wieder H, Weber WA, Becker K, Buck AK, Dobritz M, Fink U, Ulm K. Early metabolic response evaluation by fluorine-18 fluorodeoxyglucose positron emission tomography allows in vivo testing of chemosensitivity in gastric cancer: long-term results of a prospective study. Clin Cancer Res. 2008;14:2012-2018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 110] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 77. | Herrmann K, Ott K, Buck AK, Lordick F, Wilhelm D, Souvatzoglou M, Becker K, Schuster T, Wester HJ, Siewert JR. Imaging gastric cancer with PET and the radiotracers 18F-FLT and 18F-FDG: a comparative analysis. J Nucl Med. 2007;48:1945-1950. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 88] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 78. | Ott K, Herrmann K, Schuster T, Langer R, Becker K, Wieder HA, Wester HJ, Siewert JR, zum Büschenfelde CM, Buck AK. Molecular imaging of proliferation and glucose utilization: utility for monitoring response and prognosis after neoadjuvant therapy in locally advanced gastric cancer. Ann Surg Oncol. 2011;18:3316-3323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 79. | Herrmann K, Walch A, Balluff B, Tänzer M, Höfler H, Krause BJ, Schwaiger M, Friess H, Schmid RM, Ebert MP. Proteomic and metabolic prediction of response to therapy in gastrointestinal cancers. Nat Clin Pract Gastroenterol Hepatol. 2009;6:170-183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |