Published online Feb 21, 2012. doi: 10.3748/wjg.v18.i7.712
Revised: July 14, 2011
Accepted: July 21, 2011
Published online: February 21, 2012
AIM: To explore the expression pattern of OCT4 in human esophageal squamous cell carcinoma and its significance in diagnosis and prognosis.
METHODS: Using real-time polymerase chain reaction (PCR), Western blotting, immunocytochemistry and immunohistochemistry, the expression of OCT4 in three esophageal squamous cancer cell lines, KYSE70, KYSE140 and KYSE450, was characterized. OCT4 expression was investigated in a series of 153 esophageal squamous cell carcinoma samples using immunohistochemistry and explored its association with clinicopathological features.
RESULTS: Immunohistochemically, OCT4 positive immunostaining was observed in cancer cell nuclei. OCT4 was variably expressed in three esophageal squamous cancer cell lines. Among 153 specimens, 105 (68.7%) were negative or weakly positive for OCT4 staining; 21 (13.7%) were moderately positive and 27 (17.6%) were strongly positive. Higher expression level of OCT4 was significantly associated with higher histological grade (P < 0.001) and poor clinic outcome (P < 0.001).
CONCLUSION: The expression of OCT4 enables the tumor to have a higher degree of stemness, which in turn results in a poorer clinical outcome for patients with esophageal squamous cell carcinoma.
- Citation: He W, Li K, Wang F, Qin YR, Fan QX. Expression of OCT4 in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. World J Gastroenterol 2012; 18(7): 712-719
- URL: https://www.wjgnet.com/1007-9327/full/v18/i7/712.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i7.712
OCT4, also known as OCT3, belongs to the POU (Pit-Oct-Unc) transcription factor family[1]. The POU family of transcription factors can activate the expression of their target genes through binding the octameric sequence motif with an AGTCAAAT consensus sequence[2,3]. The expression of this gene is necessary for the maintenance of pluripotentiality in embryonic stem cells (ESCs) and primordial germ cells and is down-regulated in all differentiated cells in vitro as well as in vivo[2].
Previous studies have demonstrated that many cancers express OCT4 and that its expression appears to be important for cancer cell survival. Hattab et al[4] reported OCT4 immunoreactivity in all of 25 primary intracranial germinomas. Jin et al[5] discovered that the human breast cancer cell line, MCF7, expressed at least four POU gene products including OCT4. Sung et al[6] proposed that with its superior sensitivity and easy interpretation compared with other markers, OCT4 immunostaining is a powerful tool for confirming the diagnosis of retroperitoneal seminoma. In addition, OCT4 has been shown to be expressed in human tumors including pancreatic and gastric carcinomas[7,8].
Recent studies have argued that the transcription factor, OCT4, exhibited the hallmark of global regulators during mammalian embryogenesis. OCT4 works together with SOX2, another type of transcription factor, which plays important roles in the regulation of organ development and cell type specification[9,10] during embryogenesis to co-ordinate their own transcriptions[11,12], via the OCT4/SOX2 complex in ESCs[13]. OCT4 constitutes part of an important gene regulatory network and is essential for embryogenesis and/or the pluripotency and self-renewal of ESCs[14].
Cancer cells, especially in poorly differentiated or undifferentiated tumors, have been characterized by many phenotypic traits similar to undifferentiated embryonic cells[15-17]. These similarities suggest the expression of genes determining cell renewal and stemness.
Esophageal cancer is one of the leading causes of cancer-related death worldwide, especially in some high risk populations in China, such as in Linxian (Western Anyang, Henan Province). In the present study, we first attempted to characterize the expression status of OCT4 in three esophageal cell lines, and then explore their clinicopathological and survival correlations in a series of 153 esophageal cancers in patients from Anyang, Henan, China. It was found that OCT4 was expressed in all three esophageal squamous cancer cell lines (KYSE70, KYSE140 and KYSE450). Furthermore, the expression of OCT4 was significantly associated with higher histological grade and poorer clinical survival, indicating that this embryonic self-renewal factor may exert a positive influence on tumor cell stemness, which in turn results in a negative clinical course for the tumors.
In this retrospective study, 153 consecutive esophageal cancer patients who underwent potentially curative surgery without preoperative chemotherapy or radiotherapy during the period of 1990-1994 at the Anyang Tumor Hospital, Henan, China were randomly selected. Among them, 93 were men and 60 were women, ranging from 33-73 years of age with a mean age of 56.4 years. According to the International Union Against Cancer (UICC) 1997 standard, 100 were classified as stage I or II and 53 cases as stage III or IV. The patients’ information and tumor parameters are listed in Table 1. All the patients were followed at the Anyang Tumor Hospital until May 2004 supported by an international collaboration project between Anyang Tumor Hospital and the Norwegian Radium Hospital[18]. Among these patients, 97 (63.4%) patients died during the follow-up period. Surgically removed specimens were routinely fixed in buffered formalin and embedded in paraffin blocks for clinical diagnosis and reclassification for this study.
| Parameters | Cases | OCT4 | P value | ||
| Staining scores | |||||
| 0 | 1 | 2 | |||
| Age | 0.493 | ||||
| < 51 | 48 | 30 | 6 | 12 | |
| 51-60 | 52 | 39 | 6 | 7 | |
| > 60 | 53 | 36 | 9 | 8 | |
| Gender | 0.088 | ||||
| Male | 93 | 70 | 10 | 13 | |
| Female | 60 | 35 | 11 | 14 | |
| Location | 0.716 | ||||
| Upper | 14 | 10 | 2 | 2 | |
| Middle | 101 | 71 | 11 | 19 | |
| Lower | 35 | 23 | 7 | 5 | |
| Missing | 3 | ||||
| Size | 0.175 | ||||
| < 31 mm | 25 | 22 | 2 | 1 | |
| 31-60 mm | 105 | 67 | 17 | 21 | |
| > 60 mm | 14 | 10 | 1 | 3 | |
| Missing | 9 | 6 | 1 | 2 | |
| Lymph node metastasis | 0.296 | ||||
| - | 99 | 72 | 11 | 16 | |
| + | 54 | 33 | 10 | 11 | |
| UICC stage | 0.276 | ||||
| I + II | 100 | 73 | 12 | 15 | |
| III + IV | 53 | 32 | 9 | 12 | |
| T stage | 0.407 | ||||
| I | 5 | 2 | 1 | 2 | |
| II | 39 | 29 | 6 | 4 | |
| III | 100 | 68 | 14 | 18 | |
| IV | 9 | 6 | 0 | 3 | |
| Histological grade | < 0.001a | ||||
| Well | 53 | 48 | 5 | 0 | |
| Moderate | 60 | 48 | 3 | 9 | |
| Poor | 40 | 9 | 13 | 18 | |
| Patients at follow-up | 0.033a | ||||
| Alive | 56 | 45 | 3 | 8 | |
| Dead | 97 | 60 | 18 | 19 | |
The human esophageal squamous cell carcinoma cell lines, KYSE70, KYSE140 and KYSE450 (DSMZ, Germany), were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum at 37 °C under 5% CO2 and saturated moisture. The sample cells were collected and counted by trypan blue exclusion using a hemocytometer.
Immunocytochemistry was performed on all three cell lines. Simply, the cultured cells were detached and the cell suspension was centrifuged at 2000 rpm for 10 min. The supernatant was removed before the sediment was mixed with 2-3 drops of plasma and 2 drops of thrombin for 1 min, followed by the addition of 4% buffered formalin to the coagulated cell mass, which was then fixed for 30 min and placed in linen paper for further conventional paraffin cytoblock preparation. Sections (4 μm thick) were cut from the paraffin-embedded cytoblocks and immunostained according to the same procedure as the tissue blocks described below.
Multitissue array blocks were made with the MTA-1 manual tissue arrayer (Beecher Instruments Inc., Sun Prairie, WI, United States). Briefly, 4 μm sections from the routinely made paraffin blocks were stained with H and E and reevaluated to confirm the diagnosis and to identify two representative tumor areas and one stromal area. Then, the related paraffin blocks were oriented and marked. From these blocks, tissue cores with a diameter of 0.6 mm were punched and arrayed in triplicate on a recipient paraffin block. When the block construction was complete, the block was placed into a 40 °C oven overnight to tighten the cylinders by slightly melting the paraffin. Then, 5 μm sections of these tissue array blocks were cut and placed on charged Super-Frost Plus glass slides and dried in a 60 °C oven for 2-4 h. These sections were used for immunohistochemical analysis. For those samples whose tissue array materials were not representative or not available, the paraffin-embedded conventional sections were also used for additional immunohistochemistry analyses.
Immunohistochemical analysis of OCT4 was performed on 5 μm sections which were prepared from the tissue microarray blocks. The Envision Plus detection system (Dako, Carpinteria, CA, United States) was used for immunostaining. The sections were deparaffinized in xylene and microwaved in 10 mmol/L citrate buffer (pH 6.0) to unmask the epitopes. Endogenous peroxidase activity was blocked by incubation with 0.03% hydrogen peroxide in methanol for 5 min. For the detection of OCT4, the sections were incubated with the polyclonal goat anti-human OCT4 antibody (microwaving retrieval in citrate buffer in 1:40 concentration, catalog no. AF1759, R and D Systems) for 30 min at room temperature. Then, mouse anti-goat IgG (sc-2489, Santa Cruz Biotechnology Inc, Santa Cruz, CA, United States) diluted at 1:100 was added for incubation for another 30 min followed by a gentle rinse with washing buffer three times. Thereafter, the sections were incubated with a peroxidase labeled polymer conjugated to goat anti-mouse IgG (Dako, Carpinteria, CA, United States) for 30 min before staining for 5 min with 3’3-diaminobenzidine tetrahydrochloride (DAB), counterstained by hematoxylin, dehydrated and mounted in Diatex. Known OCT4 positive seminoma was used as a positive control, while the same concentration of non-immune goat IgG was applied as a negative control for OCT4.
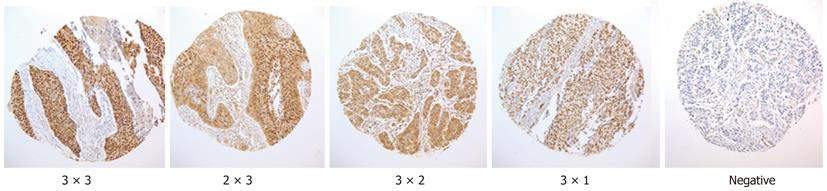
Only nuclear staining was considered as OCT4 positive. Both intensity and percentage of immunostained carcinoma cells were taken into consideration according to a previously published method with modification[19]. Briefly, the extent of positivity was scored as 0 when no positive cells were observed; 1 when the percentage of positive cells was < 10%; 2 when it was 10%-50%; and 3 when it was > 50%. The intensity was scored as 0 when no positive cells were identified; 1, weak; 2, moderate; and 3, strong staining. Multiplying the extent by intensity gave the following immunohistochemical staining grades as 0, 1, 2, 3, 4, 6 and 9. For statistical analyses, grades 0, 1 and 2 were considered as not or weakly stained and scored as 0, grades 3 and 4 were considered as moderately stained and scored as 1, and grades 6 and 9 were considered as strongly stained and scored as 2.
Total RNA and protein isolation was performed using a Total RNA and Protein Isolation kit (Macherey-Nagel, Düren, Germany) according to the user manual. About 5 × 106 cultured cells were collected and lysed. Through the NucleoSpin RNA/Protein column, RNA and DNA were bound to the column and protein was contained in the flow-through. After digestion of DNA, total RNA was isolated by washing the column. Protein was isolated from the flow-through and incubated for 3 min at 98 °C for dissolving and denaturation and stored at -20 °C until used. All of the preparations and handling steps of RNA took place in a laminar flow hood, under RNase-free conditions.
RNA quality and quantity were determined by absorbance readings at 260 nm and 280 nm with the Nano Drop (ND-1000) spectrophotometer (Wilmington, DE, United States). RNA integrity was tested by polymerase chain reaction (PCR) amplification of the gapdh. Reverse transcription of RNA was performed using Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany). cDNA was synthesized from 5 μg of total RNA isolated from the cell lines according to the manufacturer’s handbook.
The primer pairs and hydrolysis probes for quantitative real-time PCR of oct4 and gapdh were designed by Universal ProbeLibrary Assay Design Center (Roche, Mannheim, Germany). For oct4, the sense primer sequence was 5’-AGCAAAACCCGGAGGAGT-3’ and the antisense primer sequence was 5’-CCACATCGGCCTGTGTATATC-3’, giving a product of 114pb. For gapdh, the sense primer sequence was 5’-AGCCACATCGCTCAGACA-3’ and the antisense primer sequence was 5’-GCCCAATACGACCAAATCC-3’, giving a product of 66bp. The hydrolysis probes (#35, cat No. 04687680001 for oct4, and #60, cat No. 04688589001, for gapdh) were bought from Roche Diagnostics (Mannheim, Germany). In addition to the primer pairs for real-time PCR, the specific primer pair for oct4 which had been verified avoiding pseudogenes amplification by Suo et al[20] was also applied in this study for conventional PCR analyses.
Quantitative real-time PCR was performed with the LightCycler 2.0 Real-Time PCR System (Roche, Mannheim, Germany) in a total volume of 20 μL in glass-capillaries containing 2 μL of cDNA, 0.5 μmol/L of each primer, 0.1 μmol/L of hydrolysis probe and 4 μL of LightCycler TaqMan Master Mix (Roche, Mannheim, Germany). Quantification was carried out according to a published method with some modification[21]. After the expression ratio value for target gene (oct4) vs house keeping gene (gapdh) was obtained, the ratio of KYSE450 was set as 1, and all other ratio values were normalized by this value as relative quantities. The PCR was initiated with a 12 min denaturation at 95 °C and terminated with a 30 s cooling step at 40 °C. The cycling protocol consisted of denaturation at 95 °C for 10 s, annealing at 54 °C for 10 s and extension at 72 °C for 10 s and was cycled 45 times. Fluorescence detection was performed at the end of each extension step. The housekeeping gene gapdh and DEPC-H2O were set as the internal control and negative control, respectively.
The specific conventional PCR for oct4 was initiated with a 5 min denaturation at 95 °C. Amplification was carried out for 30 cycles consisting of 30 s at 95 °C, 50 s at 55 °C and 50 s at 72 °C. An additional extension step of 5 min at 72 °C was added at the end of the cycles. gapdh was used as an internal control to confirm the success of the reverse-transcription action and the identical quantity of different cDNA templates. PCR products were analyzed by electrophoresis on 1.5% agarose gel.
20 μL of protein samples were separated on a 10% SDS-acrylamide gel (Bio-Rad) for 1 h at 150V and the proteins were transferred to a nitrocellulose membrane (Whatman, Kent, United Kingdom). After blocking in 5% fat-free milk, the membrane was treated with the dilution of the primary antibody overnight at 4 °C and the dilution of the secondary IgG-horseradish peroxidase (HRP) conjugated antibody for 1 h at room temperature. The antibody used for immunohistochemistry was applied for Western blotting. The antibody was diluted in phosphate buffered saline containing 5% Blotto and 0.1% Tween-20. The stained membranes were visualized by an enhanced chemiluminescence reaction using the ECL Plus (GE Healthcare, Fairfield, CT, United States). Western blotting experiments were repeated at least three times on every sample with similar results.
Bivariate association between ordinal variables was assessed using Spearman’s correlation (exact version). For categorical data, Pearson’s χ2 test was used. All tests of statistical significance were two-sided. Overall survival was calculated from the date of diagnosis to the date of death or May 1st, 2004. Survival curves were plotted according to the Kaplan-Meier method, and the log-rank test was used to determine significant differences among groups. Multivariate analysis according to Cox’s proportional hazards regression model adjusted for clinicopathological factors (age, gender, tumor location, tumor size, lymph node metastasis, histological grade, and T stage) was performed to assess which tumor variables were independently correlated with overall survival. Statistical analyses were performed using the SPSS 16.0 package and P < 0.05 was considered as statistically significant.
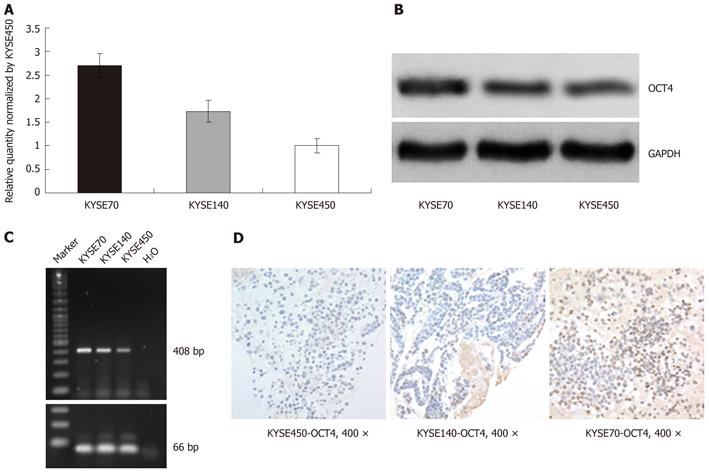
Quantitative real-time PCR was employed to analyze oct4 mRNA level in three human esophageal squamous cell carcinoma cell lines (KYSE70, KYSE140 and KYSE450). Comparatively, the ratios of oct4 vs gapdh in KYSE70 and KYSE140 were 2.7 and 1.73 times that in KYSE450, respectively (Figure 1A). To further examine the protein level of OCT4, Western blotting analysis was performed on these three cell lines. Figure 1B shows 38KD OCT4 bands, which are in agreement with the NP_002692 OCT4 characterization on NCBI. The band of OCT4 in KYSE450 was weak, but in KYSE140 and KYSE70, it was strong and stronger, respectively. Immunocytochemistry of the cell lines cytoblock paraffin sections gave similar results as shown by RT-PCR and Western blotting (Figure 1D).
Six oct4 pseudogenes have been proposed to exist using a bioinformatics approach to analyze the genomic nucleotide sequences[22]. To confirm the specific transcription of oct4, a specific primer pair for oct4 conventional PCR examination was designed and the specificity of this pair was verified by Suo et al[20]. This primer pair was also applied in the present study for additional verification of OCT4 expression. As shown in Figure 1C, the predicted PCR products (408 bp) with the specific primer pair were detected in these cell lines, highly in KYSE70 and KYSE140 and weakly in KYSE450. This verified the immunocytochemistry and Western blotting results that OCT4 was variably expressed in the human esophageal squamous cell carcinoma cell lines.
Immunohistochemically, OCT4 positive immunostaining was observed in cancer cell nuclei (Figure 2). Among the 153 tumors, 100 were negative for OCT4 staining, and 5 were scored as 1 or 2. These tumors were again classified as either negative or weakly positive for OCT4 immunostaining (105 in total, 68.7%). Twenty-one (13.7%) were scored as 3 or 4 and classified as moderately positive. Twenty-seven (17.6%) were scored as 6 or 9 and classified as strongly positive (Table 2).
| Score | OCT4 | |
| n | % | |
| 0 | 100 | 65.4 |
| 1 | 2 | 1.3 |
| 2 | 3 | 2 |
| 3 | 15 | 9.8 |
| 4 | 6 | 3.9 |
| 6 | 17 | 11.1 |
| 9 | 10 | 6.5 |
| Total | 153 | 100 |
The correlations between the clinicopathological features and OCT4 expression in the primary tumors are summarized in Table 1. The expression level of OCT4 was not correlated with age, sex, tumor location, tumor size or clinical stage. However, a higher level of OCT4 expression was significantly associated with higher histological grade (P < 0.001), indicating its correlation with dedifferentiation of these tumors.
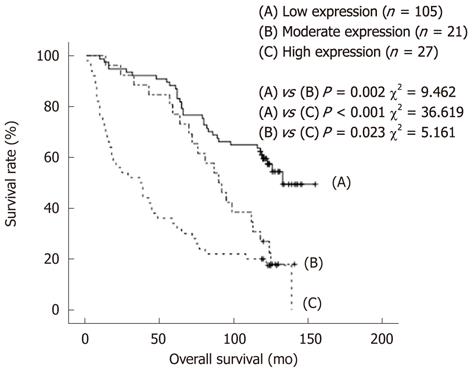
Follow-up information was available for 153 patients for a minimum period of 10 years. The median follow-up time for the 56 patients still alive was 124 mo (range 118-155 mo) and for the remaining 97 patients who died during the follow-up period was 61 mo (range 1-139 mo). In univariate analysis, patients with low OCT4 expression level in tumors had a better overall survival than patients with tumor showing moderate or high OCT4 expression level (P = 0.002 and P < 0.001, respectively, Figure 3).
In multivariate Cox’s regression analysis, we detected a 2.625-fold increased risk of tumor-related death for patients (P < 0.001) whose tumors showed high OCT4 expression level compared to patients with low expression level (Table 3).
The expression of OCT4 was first discovered in human esophageal squamous cancer cell lines with the antibody AF1759 from R and D System for immunocytochemistry in the authors’ lab. Since controversial results concerning the expression of OCT4 in tumors exist[23], the expression status of this factor in these cell lines was investigated. The expressions were repeatedly demonstrated immunocytochemically, with different positive and negative controls. To verify these findings, real-time PCR with hydrolysis probes was carried out, and repeatedly demonstrated similar results in these cell lines. Considering the fact that oct4 pseudogenes are also transcribed[20], the oct4 specific PCR primer pair (poct4a, 5’-TCCCTTCGCAAGCCCTCAT-3’ and poct4b, 5’-TGACGGTGCAGGGCTCCGGGGAGGCCCCATC-3’) were applied in conventional PCR for these three cell lines, obtaining similar results. All these experiments were repeated at least twice. The additional Western blotting analysis also disclosed similar results as shown by immunocytochemistry and PCR. Therefore, we can conclude that OCT4 is variably expressed in these esophageal squamous cancer cell lines.
Based on the findings on cell lines, this work was extended to clinical esophageal cancer samples for which official follow-up data were obtained during the international collaboration. To date, this study is the first to report the expression patterns of the embryonic stem cell factor OCT4 in esophageal squamous carcinomas.
Esophageal cancer is one of the most aggressive neoplasms and the overall prognosis for esophageal cancer patients is poor[24]. One of the reasons for the low survival rate is the tumor’s intrinsic resistance to many clinical therapies, especially chemotherapy. Chemotherapy often removes the bulk of a tumor mass without preventing tumor recurrence, suggesting the survival of a subset of cancer stem cells. Recent studies have provided experimental evidence for the concept that human tumor growth may depend on a small portion of cancer stem cells[25].
The present investigation on esophageal squamous carcinomas revealed several novel observations. Firstly, it was discovered that 17.6% of these esophageal cancer samples had strong positive expression of OCT4. Secondly, although the expression level of OCT4 did not correlate with age, sex, tumor size, location, histological type or UICC stage, it was significantly associated with higher histological grade of the tumors and poorer clinical outcome of the patients.
It has been proposed that OCT4 is a key regulator of stem cell pluripotency and differentiation, indicating that it is the primary factor determining the fate of ESCs by controlling cell self-renewal and differentiation[26]. Phenotypically, human preimplantation embryonic cells resemble cancer cells in many ways, especially in their ability to grow indefinitely. Both types of cells undergo deprogramming to a proliferating state and become immortal, self-renewable and invasive. It has been reported that OCT4 is expressed in human tumors but not in normal somatic tissues[27], in agreement with the hypothesis that embryonic genes are re-activated in tumor cells.
In this series, OCT4 expression was strongly associated with the histological grade of the tumors and survival of the patients, which supports the findings of previous reports[28-30]. Given the previously reported functions of OCT4, the results suggest that it may play a role in conferring a less differentiated phenotype or inactivating the ability to differentiate in esophageal carcinomas.
It has been demonstrated that human somatic cells can be reprogrammed into pluripotent stem cells by either a combination of OCT4, SOX2, Nanog and Lin28 factors[31] or a combination of OCT4, SOX2, Klf4 and c-Myc factors[32]. A common feature of these studies is the contribution of OCT4 and SOX2 for pluripotency, indicating that tumor cells with OCT4 expression may behave as or close to tumor stem cells. It is also known that SP positive tumor cells, which most probably harvest tumor stem cells, are resistant to chemotherapy and radiotherapy[33,34]. Thus, the present investigation on esophageal squamous cancer samples may indicate that OCT4 conveys a stemness feature in tumor cells, so that these tumors could easily develop and relapse, resulting in poorer clinical outcome.
The current results demonstrate that the ESC marker, OCT4, could be detected in human esophageal squamous cancer cell lines and related cancer tissues. The poorly differentiated esophageal squamous cancer cell lines expressed higher levels of OCT4. The higher expression level of OCT4 in esophageal carcinomas was correlated with higher histological grade and poorer overall survival of esophageal squamous cell cancer patients. Since the function of pluripotency and self-renewal of this factor has been characterized in adult human cells[31,32], the role of OCT4 in esophageal carcinogenesis, especially in consideration of tumor cell stemness, merits further studies.
The embryonic stem cell factor, OCT4, is essential for pluripotency and self-renewal of embryonic stem cells. Cancer cells, especially in poorly differentiated or undifferentiated tumors, have been characterized by many phenotypic traits similar to undifferentiated embryonic cells, indicating that OCT4 may be expressed in solid tumors.
Previous studies have demonstrated that many cancers express OCT4 and that its expression appears to be important for cancer cell survival, such as in germinoma, breast cancer, seminoma, pancreatic and gastric carcinomas. However, the expression pattern of OCT4 in human esophageal squamous cell carcinoma and its significance in diagnosis and prognosis remain unclear. In this study, the authors demonstrated that OCT4 positive immunostaining was observed in cancer cell nuclei, and the expression of OCT4 enabled the tumor to have a higher degree of stemness, which in turn resulted in poorer clinical outcome for the patients with esophageal squamous cell carcinomas.
To date, this study is the first to report the expression patterns of the embryonic stem cell factor, OCT4, in esophageal squamous carcinomas. The present investigation revealed several novel observations. Firstly, it was discovered that 17.6% of these esophageal cancer samples had strong positive expression for OCT4. Secondly, although the expression level of OCT4 did not correlate with age, sex, tumor size, location, histological type or International Union Against Cancer stage, it was significantly associated with higher histological grade of the tumors and poorer clinical outcome of the patients.
By knowing that the embryonic stem cell (ESC) marker, OCT4, can be detected in human esophageal squamous cancer cell lines and related cancer tissues, and the higher expression level of OCT4 in esophageal squamous carcinomas was correlated with higher histological grade and poorer overall survival of esophageal squamous cell cancer patients, this study may represent a future strategy for therapeutic intervention in the treatment of patients with esophageal squamous carcinoma.
OCT4 belongs to the POU (Pit-Oct-Unc) transcription factor family. The expression of this gene is necessary for the maintenance of pluripotentiality in ESCs and primordial germ cells and is down-regulated in all differentiated cells in vitro as well as in vivo.
This study considers the investigation of the expression of the embryonic stem cell factor OCT4 in three esophageal cell lines and additionally in esophageal squamous carcinoma patients. Important finding of this study was the observation that OCT4 expression was strongly associated with the histological grade of the tumors and survival of the patients. The poorly differentiated esophageal squamous cancer cell lines expressed higher level of OCT4. This study make a great contribution to studies of OCT4 as one of the transcription factors and its role in cancer cell’s survival. The study support the hypothesis that the expression of OCT4 as embryonic self-renewal factor may exert a positive influence on tumor cell stemness, which in turn results in a negative clinical course for the tumors.
Peer reviewer: Tamara Vorobjova, MD, PhD, Dr., Scimed. Senior Researcher in Immunology, Department of Immunology, Institute of General and Molecular Pathology, University of Tartu, Ravila, 19, Tartu 51014, Estonia
S- Editor Sun H L- Editor Webster JR E- Editor Zhang DN
| 1. | Schöler HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435-439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 540] [Cited by in F6Publishing: 561] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 2. | Pesce M, Schöler HR. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271-278. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 589] [Cited by in F6Publishing: 578] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 3. | Schöler HR. Octamania: the POU factors in murine development. Trends Genet. 1991;7:323-329. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 246] [Cited by in F6Publishing: 285] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 4. | Hattab EM, Tu PH, Wilson JD, Cheng L. OCT4 immunohistochemistry is superior to placental alkaline phosphatase (PLAP) in the diagnosis of central nervous system germinoma. Am J Surg Pathol. 2005;29:368-371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 89] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 5. | Jin T, Branch DR, Zhang X, Qi S, Youngson B, Goss PE. Examination of POU homeobox gene expression in human breast cancer cells. Int J Cancer. 1999;81:104-112. [PubMed] [Cited in This Article: ] |
| 6. | Sung MT, MacLennan GT, Cheng L. Retroperitoneal seminoma in limited biopsies: morphologic criteria and immunohistochemical findings in 30 cases. Am J Surg Pathol. 2006;30:766-773. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409-1420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 642] [Cited by in F6Publishing: 659] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 8. | Tsukamoto T, Mizoshita T, Mihara M, Tanaka H, Takenaka Y, Yamamura Y, Nakamura S, Ushijima T, Tatematsu M. Sox2 expression in human stomach adenocarcinomas with gastric and gastric-and-intestinal-mixed phenotypes. Histopathology. 2005;46:649-658. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 82] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 9. | Jin T, Branch DR, Zhang X, Qi S, Youngson B, Goss PE. Examination of POU homeobox gene expression in human breast cancer cells. Int J Cancer. 1999;81:104-112. [PubMed] [Cited in This Article: ] |
| 10. | Boiani M, Schöler HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005;6:872-884. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 501] [Cited by in F6Publishing: 488] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 11. | Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J Biol Chem. 2005;280:5307-5317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 276] [Cited by in F6Publishing: 277] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 12. | Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 2002;30:3202-3213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 235] [Cited by in F6Publishing: 251] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 13. | Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031-6046. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 14. | Iki K, Pour PM. Expression of Oct4, a stem cell marker, in the hamster pancreatic cancer model. Pancreatology. 2006;6:406-413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Markert CL. Neoplasia: a disease of cell differentiation. Cancer Res. 1968;28:1908-1914. [PubMed] [Cited in This Article: ] |
| 16. | Potter VR. Phenotypic diversity in experimental hepatomas: the concept of partially blocked ontogeny. The 10th Walter Hubert Lecture. Br J Cancer. 1978;38:1-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 198] [Cited by in F6Publishing: 216] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Till JE. Stem cells in differentiation and neoplasia. J Cell Physiol Suppl. 1982;1:3-11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 69] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Yang H, Berner A, Mei Q, Giercksky KE, Warloe T, Yang G, Cui J, Suo Z, Zhang S, Nesland JM. Cytologic screening for esophageal cancer in a high-risk population in Anyang County, China. Acta Cytol. 2002;46:445-452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Liu FS, Hsieh YT, Chen JT, Ho ES, Hung MJ, Lin AJ. FHIT (fragile histidine triad) gene analysis in cervical intraepithelial neoplasia. Gynecol Oncol. 2001;82:283-290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | Suo G, Han J, Wang X, Zhang J, Zhao Y, Zhao Y, Dai J. Oct4 pseudogenes are transcribed in cancers. Biochem Biophys Res Commun. 2005;337:1047-1051. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 154] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 21. | Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24144] [Cited by in F6Publishing: 24572] [Article Influence: 1068.3] [Reference Citation Analysis (0)] |
| 22. | Pain D, Chirn GW, Strassel C, Kemp DM. Multiple retropseudogenes from pluripotent cell-specific gene expression indicates a potential signature for novel gene identification. J Biol Chem. 2005;280:6265-6268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 95] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 23. | Cantz T, Key G, Bleidissel M, Gentile L, Han DW, Brenne A, Schöler HR. Absence of OCT4 expression in somatic tumor cell lines. Stem Cells. 2008;26:692-697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 96] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 24. | Lin YC, Wu MY, Li DR, Wu XY, Zheng RM. Prognostic and clinicopathological features of E-cadherin, alpha-catenin, beta-catenin, gamma-catenin and cyclin D1 expression in human esophageal squamous cell carcinoma. World J Gastroenterol. 2004;10:3235-3239. [PubMed] [Cited in This Article: ] |
| 25. | Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6844] [Cited by in F6Publishing: 6706] [Article Influence: 291.6] [Reference Citation Analysis (0)] |
| 26. | Pan GJ, Chang ZY, Schöler HR, Pei D. Stem cell pluripotency and transcription factor Oct4. Cell Res. 2002;12:321-329. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 236] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 27. | Monk M, Holding C. Human embryonic genes re-expressed in cancer cells. Oncogene. 2001;20:8085-8091. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 254] [Cited by in F6Publishing: 260] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 28. | Sanada Y, Yoshida K, Ohara M, Oeda M, Konishi K, Tsutani Y. Histopathologic evaluation of stepwise progression of pancreatic carcinoma with immunohistochemical analysis of gastric epithelial transcription factor SOX2: comparison of expression patterns between invasive components and cancerous or nonneoplastic intraductal components. Pancreas. 2006;32:164-170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 107] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 29. | Ruangpratheep C, Lohachittranond C, Poonpracha T, Punyarit P. OCT4 expression on a case of poorly differentiated (insular) carcinoma of the thyroid gland and minireview. J Med Assoc Thai. 2005;88 Suppl 3:S281-S289. [PubMed] [Cited in This Article: ] |
| 30. | Spisek R, Kukreja A, Chen LC, Matthews P, Mazumder A, Vesole D, Jagannath S, Zebroski HA, Simpson AJ, Ritter G. Frequent and specific immunity to the embryonal stem cell-associated antigen SOX2 in patients with monoclonal gammopathy. J Exp Med. 2007;204:831-840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 152] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 31. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7589] [Cited by in F6Publishing: 6999] [Article Influence: 411.7] [Reference Citation Analysis (0)] |
| 32. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14327] [Cited by in F6Publishing: 13579] [Article Influence: 848.7] [Reference Citation Analysis (0)] |
| 33. | Bunting KD. ABC transporters as phenotypic markers and functional regulators of stem cells. Stem Cells. 2002;20:11-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 331] [Cited by in F6Publishing: 356] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 34. | Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, Goodell MA, Brenner MK. A distinct "side population" of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci USA. 2004;101:14228-14233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 977] [Cited by in F6Publishing: 1009] [Article Influence: 50.5] [Reference Citation Analysis (0)] |