Published online Dec 21, 2012. doi: 10.3748/wjg.v18.i47.7003
Revised: May 31, 2012
Accepted: June 8, 2012
Published online: December 21, 2012
AIM: To investigate the association between interleukin-28B (IL28B) genotype and response to treatment and hepatic fibrosis in patients with hepatitis C virus (HCV) genotype 4.
METHODS: Two hundred and one HCV-genotype 4 patients were included. All patients were treated with Peginterferon alph2a/Ribavirin for 48 wk. End of treatment response (ETR) was defined as loss of detectable serum HCV RNA at the end of treatment. Sustained viral response (SVR) was defined as loss of detectable serum HCV RNA at the end of 24 wk follow up. Genotyping of IL28B rs12979860 was performed using the TaqMan assay. We used logistic regression to estimate the adjusted odds ratio (aOR) and 95%CI.
RESULTS: The study included 201 HCV-genotype 4 patients. The majority of patients were men (89.6%), with a median age of 47 years, inter-quartile range (40-51). Approximately 62.5% of patients had ETR, and 49.6% had SVR. Individuals who achieved SVR were more likely to be younger (χ2 = 4.91, P = 0.027), and less likely to have fibrosis (χ2 = 15.54, P < 0.0001), or inflammation (χ2 = 7.58, P = 0.006). The genotype distribution of rs12979860 was 36.2%, 49.0% and 14.8% for genotypes CC, CT, and TT, respectively. In these participants, rs12979860 genotype distribution did not differ by gender (P = 0.466), pretreatment viral load (P = 0.600), inflammation (P = 0.435), or fibrosis (P = 0.291). The frequencies of IL28B rs12979860 genotypes were TT (14.8%), CT (49.0%), and CC (36.2%). Compared to rs12979860 genotype TT, aORs (95%CI) for ETR and SVR were: CC genotype, [17.55 (5.34-57.69) and 5.92 (2.09-16.76), respectively]; CT genotype, [5.15 (1.80-14.78) and 2.48 (0.94-6.52), respectively]. In the current study, the patients who did not achieve ETR or SVR had a lower prevalence of rs12979860 CC (17.4% and 23.3%, respectively) than individuals who had ETR or SVR (47.9% and 47.2%, respectively). Individuals with rs12979860 CC genotype had approximately 6 times the odds of SVR compared to individuals with TT genotype (aOR = 5.92; 95%CI: 2.09-16.76). Similarly, patients with CT genotype had SVR more often than patients with TT genotype (aOR = 2.48; 95%CI: 0.94-6.52). Carrying at least one copy of the C allele (genotypes CT and CC) had almost 8 times the probability of ETR compared to those with genotype rs12979860 TT (aOR = 7.87; 95%CI: 2.84-21.82), and approximately 3 times the odds of SVR compared to those with genotype rs12979860 TT (aOR = 3.46; 95%CI: 1.37-8.74). In addition, data were consistent with a significant gene-dose relationship (aOR = 4.05/allele; 95%CI: 2.27-7.22). The association between rs12979860 genotype and SVR was similar among those who achieved and those who did not achieve SVR.
CONCLUSION: In HCV-genotype 4 patients, rs12979860 is a sensitive predictor of viral clearance, independent of viral load, age, gender or fibrosis, with no similar relation to severity of fibrosis.
-
Citation: Derbala M, Rizk N, Shebl F, Alkaabi S, Eldweik N, John A, Sharma M, Yaqoob R, Almohanadi M, Butt M, Alejji K.
Interleukin-28 and hepatitis C virus genotype-4: Treatment-induced clearance and liver fibrosis. World J Gastroenterol 2012; 18(47): 7003-7008 - URL: https://www.wjgnet.com/1007-9327/full/v18/i47/7003.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i47.7003
Hepatitis C virus genotype 4 (HCV-G4) is the most common type of HCV in the Middle East and Africa, particularly in Egypt, which has the highest prevalence of HCV worldwide (15%), and HCV-G4 represents 90% of all HCV cases[1]. Despite the development of new direct antiviral agents such as protease inhibitors, which have improved response in HCV genotypes[1,2], unfortunately G4 has emerged as a resistant genotype to new treatment strategies. This raises the importance, both for patient care and the economic approach, of improving the prediction of response to standard pegylated interferon alfa (PEG-IFN) and ribavirin therapy.
Although the mechanisms leading to clearance of acute HCV infection are not completely understood, both innate and adaptive immune responses have been suggested to play crucial roles in viral eradication and response to treatment[3]. Besides immune responses, other host factors have also been associated with treatment-induced viral clearance. Higher rates of viral resolution have been reported in women compared with men; however, other factors such as age, race, or route of transmission were not significantly associated with viral resolution[4].
Recently, another host factor, a genetic variation in the interleukin-28B (IL28B) gene, was found to predict spontaneous clearance of HCV infection[5]. A single nucleotide polymorphism (SNP) rs12979860 located 3 kb upstream of IL28B, the gene that encodes IFN-λ3, has been strongly associated with resolution of HCV infection[6]. Patients with C/C genotype were more likely to eradicate HCV than those with T/T genotype[7,8]. The same SNP has also been associated with treatment-induced viral clearance[9,10]. Patients with the C/C genotype were twice as likely to achieve an sustained viral response (SVR) compared to patients with other IL28B SNP genotypes[6]. These findings jointly suggest a role for IFN-λ in the innate control of HCV. Of note, the T/T genotype was shown to be more common among those with African ancestry[5].
However, the association between IL28B genotype and response to treatment among individuals infected with HCV-G4 still needs further investigation among the ethnic group living in the Middle East. Therefore, we conducted the present study to assess the extent of the association between IL28B genotype and response to treatment in HCV-G4 and severity of liver fibrosis.
The study included 201 Egyptian patients with chronic HCV genotype-4 who were followed in the Hamad Medical Corporation outpatient clinic in the State of Qatar. Patients received HCV treatment between 2007 and 2010. This was a retrospective-prospective cohort study in which all patients who were treated between 2007 and 2010 and had finished their follow up to week 72 were invited to participate (retrospective aspect). In addition, all patients who were currently on treatment and had not completed 72 wk of follow up (thus their outcome was not known yet) were invited to participate in the study (prospective aspect) and were followed until week 72 to determine their outcome.
All patients provided written informed consent in accordance with the Declaration of Helsinki of 1979, and the ethics research committee of the Hamad Medical Corporation provided ethical approval.
Chronic HCV infection was diagnosed by a sustained increase in alanine aminotransferase (ALT), positive anti-HCV serology and active virus replication shown by the detection of HCV-RNA and histological pattern of chronic active hepatitis. Patients were excluded from treatment if they had: active alcohol consumption over 80 g/d, concurrent hepatic B virus, immunodeficiency viruses, autoimmune hepatitis, hemochromatosis, Wilson disease, or were on antiviral or corticosteroid therapy.
All patients were treated with 180 μg of Peginterferon-2a (Pegasys®, Hoffmann-La Roche, Basel, Switzerland) subcutaneously once weekly and Ribavirin (COPEGUS®; Hoffmann-La Roche) 1000 mg (body weight ≤ 75 kg) or 1200 mg (body weight ≥ 75 mg) orally for 48 wk. End of treatment response (ETR) was defined as loss of detectable serum HCV RNA at the end of treatment (48 wk). SVR was defined as loss of detectable serum HCV RNA at the end of follow up (72 wk).
Viral assays: Testing for anti-HCV was carried out using a commercial ELISA kit (Axsym 3.0; Abbott Laboratories, Chicago, IL, United States). All patients were HCV-G4 as detected by the Inno-LiPA HCV II assay (Innogenetics Inc., Alpharetta, GA, United States). Serum HCV RNA level monitoring was by Amplicor (version 2.0; Hoffmann-La Roche) with a minimum detection limit of 50 IU/mL.
Liver histology: The necro-inflammatory and fibrosis scores were assigned based on the Scheuer scoring system from 0 to 4. The patients were further subdivided into mild fibrosis (stages I and II) and severe fibrosis (stages III and IV).
IL28B genotype assay: Genomic DNA was extracted from EDTA whole-blood samples using the QiaAmp DNA Blood Mini Kit # 51166 (Qiagen GmbH, Hilden, Germany). DNA concentration was measured using a Nanodrop Spectrophotometer to assess the quantity of the product. Polymorphisms of the studied SNP were carried out by the 5’ nuclease assay using the TaqMan MGB probe. The reaction was performed using an ABI 7500 (Applied Biosystems, Foster City, CA, United States) in the Biomedical Labs-Health Sciences Department at Qatar University, Doha, Qatar. The primers and the TaqMan MGB probes of the SNP were provided by the Custom assay-on demandTM service (Applied Biosystems). The 5’ nuclease assay was performed using 10-30 ng genomic DNA, 1 × TaqMan Universal polymerase chain reaction Master Mix (Applied Biosystems), and 1 × primer/probe mix using the correct conditions for amplifications according to the manufacturer’s instructions. Negative controls as well as non-template controls were included in each run.
In the bivariate analyses we used the χ2 test for categorical variables and ANOVA for continuous variables to compare the relevant characteristics by treatment response status. We conducted multivariable logistic regression to calculate adjusted odds ratio (aOR) and 95%CI of the association between treatment response and IL28B genotype. Only variables that were significantly associated with treatment response or IL28B genotype were included in the multivariable models. To test whether the association between IL28 genotype and response to treatment was modified by gender, grade, viral load, inflammation or fibrosis, we employed logistic regression models with an interaction term (cross product) for IL28 genotype and the modifier of interest included. ORs were computed using the homozygous minor allele as the reference group. All analyses were performed using SAS program version 9.2 (SAS Institute, Cary, NC, United States).
Table 1 presents the characteristics of the study population. The study included 201 patients. The majority of participants were males (89.6%). Median age was 47 years (inter-quartile range 40-51). We observed Rapid Virological response (RVR), ETR, and SVR in 52.5%, 62.5% and 54.2% of patients, respectively. The mean, median and range of viral load (log 10) over the duration of therapy among responders and non-responders, were (4.9, 2.2, 0.0-6.9 and 5.5, 3.9, 2.1-6.9) for RVR, (3.6, 1.0, 0.0-4.23 and 6.0, 4.9, 2.5-6.5) for ETR, respectively. There was no difference in gender, or pretreatment HCV viral load by RVR, ETR or SVR status. RVR was not associated with age, liver inflammation or fibrosis. Those who had ETR were less likely to have fibrosis (χ2 = 12.54, P < 0.001), or inflammation (χ2 = 5.17, P = 0.023). Only 62.5% of patients had an ETR and 49.6% had SVR. There was no difference in gender, or pretreatment HCV viral load by ETR or SVR status. In addition, individuals who achieved SVR were more likely to be younger (χ2 = 4.91, P = 0.027), and less likely to have fibrosis (χ2 = 15.54, P < 0.0001), or inflammation (χ2 = 7.58, P = 0.006).
| End of treatment response | Sustained viral response | |||
| No | Yes | No | Yes | |
| Age, median (IQR) | 48.0 (40.5-53.0) | 46.0 (40.0-51.0) | 48.0 (43.5-53.0) | 46.0 (38.0-51.0) |
| HCV log10 viral load, median (IQR) | 5.9 (5.1-6.4) | 5.5 (3.8-6.3) | 5.8 (5.2-6.5) | 5.4 (3.6-6.1) |
| Male (%) | 93.1 | 87.5 | 91.3 | 88.1 |
| rs12979860 (%) | ||||
| TT | 30.0 | 5.1 | 23.3 | 7.6 |
| CT | 52.9 | 47.0 | 53.4 | 45.2 |
| CC | 17.1 | 47.9 | 23.3 | 47.2 |
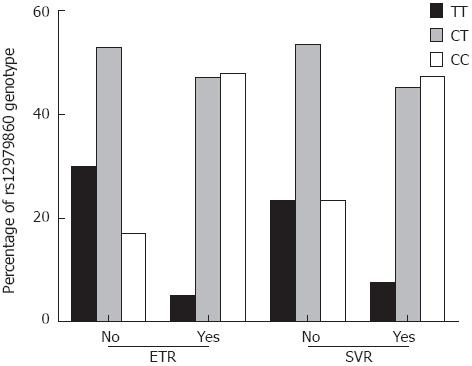
The genotype distribution of rs12979860 was 36.2%, 49.0% and 14.8% for genotypes CC, CT, and TT, respectively. Among our participants, rs12979860 genotype distribution did not differ by gender (P = 0.466), pretreatment viral load (P = 0.600), inflammation (P = 0.435), or fibrosis (P = 0.291). In our study, the patients who did not achieve ETR or SVR had a lower prevalence of rs12979860 CC (17.4% and 23.3%, respectively) than individuals who had ETR or SVR (47.9% and 47.2%, respectively). Genotype distributions by HCV status are presented in Figure 1.
The data revealed that adjusting for stage, patients with rs12979860 CC and CT genotypes had ETR more often than patients with TT genotype [aOR (95%CI) = 17.55 (5.34-57.69) and 5.15 (1.80-14.78), respectively]. Patients carrying at least one copy of the C allele (genotypes CT and CC) had almost 8 times the probability of ETR compared to those with genotype rs12979860 TT (aOR = 7.87; 95%CI: 2.84-21.82) (Table 2). In addition, data were consistent with a significant gene-dose relationship (aOR = 4.05/allele; 95%CI: 2.27-7.22). No significant interactions were detected by gender, fibrosis, inflammation, or pretreatment ALT levels (P > 0.05).
| End of treatment response | Sustained viral response | |||
| OR (95%CI) | P value | OR (95%CI) | P value | |
| rs12979860 | ||||
| TT | Ref. genotype | Ref. genotype | ||
| CT | 5.15 (1.80-14.78) | 0.002 | 2.48 (0.94-6.52) | 0.065 |
| CC | 17.55 (5.34-57.69) | < 0.0001 | 5.92 (2.09-16.76) | < 0.001 |
Individuals with rs12979860 CC genotype had approximately 6 times the odds of SVR compared to individuals with TT genotype (aOR = 5.92; 95%CI: 2.09-16.76). Similarly, patients with CT genotype had SVR more often than those with TT genotype (aOR = 2.48; 95%CI: 0.94-6.52). Carrying at least one copy of the C allele (genotypes CT and CC) had approximately 3 times the odds of SVR compared to those with genotype rs12979860 TT (aOR = 3.46; 95%CI: 1.37-8.74) (Table 2). Of note, we observed a significant gene-dose relationship (aOR = 2.42/allele; 95%CI: 1.47-3.99). We did not reveal any significant interactions by gender, fibrosis, inflammation, or pretreatment ALT levels (P > 0.05).
There were no significant interactions between the rs12979860 genotype and RVR indicating that the association between the rs12979860 genotype and SVR was similar among those who achieved RVR and those who did not achieve RVR.
Recent studies have suggested that SNPs within or adjacent to IL28B are the most promising, host-related predictors of treatment-induced clearance in HCV genotype-1 patients[11,12], however, in studies of individuals infected with genotypes 2 and 3, the association between IL28B genotype and outcome is less pronounced, including rs12979860 with treatment-induced clearance[13,14]. The impact of genetic IL28B variability on viral elimination during therapy in G4 is still limited. Similar to HCV genotype-1, in the present study, we demonstrated that CC genotypes of rs12979860 significantly determined SVR in patients with HCV G4. Not only that, even carrying at least one copy of the C allele increased sensitivity to PEG-IFN/Ribavirin therapy, which was 3 times that of rs12979860-negative hosts. The mechanism and explanation behind the association between genetic variations in the IL28B gene and spontaneous clearance may be related to the host innate immune response. IL28B encodes IFN-λ3, which is involved in viral control, including HCV. In vitro, IFN-α induces expression of IFN-λ genes, which inhibit HCV replication[15]. IL28A&b and IL29 are three closely related cytokine genes that encode proteins known as type III IFNs at chromosomal region 19q13. The three cytokines are induced by viral infection and have antiviral activity. In addition, an experimental genetic variant regulating IL28 expression is important for PEG-IFN TLR-mediated antiviral protection.
HCV treatment should not be withheld based solely on IL28B genotype, as patients with the TT genotype can achieve SVR, as it was found that the association between rs12979860 genotype and SVR was similar between the patients regardless of their RVR status.
In contrast to Ge et al[6] who reported higher viral load with the C allele, in our participants, rs12979860 genotype distribution did not differ by pretreatment viral load.
Rao et al[16] reported a significant genetic polymorphism among women who responded to treatment, but Montes-Cano et al[17] found that rs12979860 genotype distribution did not differ by gender in G4 patients. This can be explained simply, by the difference in tested SNPs, while Rao et al[16] reported this gender-related difference with the rs8099917 TT genotype, in our study and in the study by Montes-Cano, we studied rs12979860.
The association between IL28B polymorphisms and liver fibrosis progression is still controversial[18]. While, Barreiro et al[19] reported that IL28B CC carriers might experience a more rapid progression of HCV-related liver fibrosis, Marabita et al[20] reported that IL28B polymorphisms had no impact on fibrosis progression. Some investigators found that the unfavorable rs12979860 T/T gene pattern was associated with worse liver fibrosis[21], while others did not replicate this finding. In G4 patients, and similar to Asselah et al[22], no significant differences and associations for genotype and alleles frequencies were detected between mild and severe fibrosis. Host genetic factors associated with the risk of liver disease progression in hepatitis C-G4 needs further investigation.
Few reports have studied the relation between IL28B and G4, particularly among patients in our area, where Egyptians represent the majority of cases with different ethnic and G4-subtypes[23]. The overall frequency of the protective C allele in our study was 85%, which was higher than that found by Kurbanov et al[24], who reported a frequency of 67% among the Egyptians studied. When the diploid genotype was considered, the overall frequency of the protective C/C genotype was 45%, which was close to our finding (36.2%). With regard to the association between genetic polymorphisms and response to treatment, Asselah et al[22] reported that SNP rs12979860 was strongly associated with SVR in patients infected with HCV-G4 in 164 HCV-G4 patients who were from different ethnic groups (Egyptian, European, and Sub-Saharan African). Although Egyptians represented 67% (75/112) of the patients with G4 in the study by De Nicola et al[25], the results were very similar to ours concerning IL28B genotype distribution and distribution of the C allele among the patients who achieved SVR. Our study re-enforces the importance of SNP rs12979860 of IL28B in G4, regardless of viral clearance by week 4 and confirmed the absence of its correlation with hepatic fibrosis.
This study specifically examined the relationship between IL28B rs12979860 CC genotype, rapid viral clearance, treatment response and severity of liver disease in patients with chronic HCV-G4 infection which included a large sample of patients from the same ethnic group.
In conclusion, in patients with HCV-G4, the IL28B single nucleotide polymorphism (rs12979860) is a sensitive predictor of viral clearance, independent of baseline viral load, age, gender or fibrosis. This polymorphism does not have a similar relation with rapid viral clearance or severity of hepatic fibrosis.
The authors wish to thank Mrs. Mooza Al-Khinji and Ms. Randa Saleh, Biomedical Labs, CAS, at Qatar University, Qatar for their technical support in the genotyping experiments.
Genotype 4 (G4) has emerged as a resistant genotype to new treatment strategies. This raises the importance, both for patient care and the economic approach, of improving the prediction of response to standard pegylated interferon alfa and ribavirin therapy. Genetic variation in the interleukin-28B (IL28B) gene has been found to predict spontaneous clearance of hepatitis C virus (HCV) infection and response to treatment.
The association between the IL28B genotype and response to treatment in individuals infected with HCV-G4 still needs further investigation, especially among the ethnic group living in the Middle East. The research focused on the sensitivity of this gene in the prediction of response and its importance in predicting liver histology progression.
In previous reports, a few G4 patients were studied which was mainly related to spontaneous clearance. This study provided information on the relation between IL28B and post-treatment viral clearance and its relation to hepatic fibrosis in G4.
The study results suggest that the IL28B single nucleotide polymorphism (rs12979860) is a sensitive predictor of viral clearance in G4 patients and may improve clinical prediction models for treatment and should be included as a test in pre-treatment investigations, providing an opportunity for clinicians to individualize treatment regimens for hepatitis C patients.
The authors studied 201 G4 HCV infected patients. All patients were treated with Peginterferon/Ribavirin for 48 wk. They found in HCV-G4, IL28B single nucleotide polymorphism (rs12979860) is an independent predictor of viral clearance.
Peer reviewer: Dr. Jin-Town Wang, Professor, Department of Microbiology and Internal Medicine, National Taiwan University, College of Medicine, 1, Sec 1, Jen-Ai Rd, Taipei 10016, Taiwan, China
S- Editor Lv S L- Editor Webster JR E- Editor Zhang DN
| 1. | Kamal SM. Hepatitis C virus genotype 4 therapy: progress and challenges. Liver Int. 2011;31 Suppl 1:45-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 2. | Manns MP, Markova AA, Calle Serrano B, Cornberg M. Phase III results of Boceprevir in treatment naïve patients with chronic hepatitis C genotype 1. Liver Int. 2012;32 Suppl 1:27-31. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest. 2009;119:1745-1754. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 394] [Cited by in F6Publishing: 405] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 4. | Asselah T, Estrabaud E, Bieche I, Lapalus M, De Muynck S, Vidaud M, Saadoun D, Soumelis V, Marcellin P. Hepatitis C: viral and host factors associated with non-response to pegylated interferon plus ribavirin. Liver Int. 2010;30:1259-1269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 126] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 5. | Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798-801. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1667] [Cited by in F6Publishing: 1635] [Article Influence: 109.0] [Reference Citation Analysis (0)] |
| 6. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2776] [Cited by in F6Publishing: 2666] [Article Influence: 177.7] [Reference Citation Analysis (0)] |
| 7. | Shebl FM, Pfeiffer RM, Buckett D, Muchmore B, Chen S, Dotrang M, Prokunina-Olsson L, Edlin BR, O'Brien TR. IL28B rs12979860 genotype and spontaneous clearance of hepatitis C virus in a multi-ethnic cohort of injection drug users: evidence for a supra-additive association. J Infect Dis. 2011;204:1843-1847. [PubMed] [Cited in This Article: ] |
| 8. | Scott J, Holte S, Urban T, Burgess C, Coppel E, Wang C, Corey L, McHutchison J, Goldstein D. IL28B genotype effects during early treatment with peginterferon and ribavirin in difficult-to-treat hepatitis C virus infection. J Infect Dis. 2011;204:419-425. [PubMed] [Cited in This Article: ] |
| 9. | Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100-1104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1505] [Cited by in F6Publishing: 1482] [Article Influence: 98.8] [Reference Citation Analysis (0)] |
| 10. | Halfon P, Bourliere M, Ouzan D, Maor Y, Renou C, Wartelle C, Pénaranda G, Tran A, Botta D, Oules V. A single IL28B genotype SNP rs12979860 determination predicts treatment response in patients with chronic hepatitis C Genotype 1 virus. Eur J Gastroenterol Hepatol. 2011;23:931-935. [PubMed] [Cited in This Article: ] |
| 11. | Asselah T, Essioux L, Marcellin P, Fried MW, Jensen DM, Germer S, Benayed R, Chu T, Tietz A, Chin D. A chromosome 19 SNP (RS12979860) predicts outcome (EVR/SVR) in HCV patients treated with interferon, independent of pegylation or ribavirin. J Hepatol. 2010;A1180. [Cited in This Article: ] |
| 12. | Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338-1345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 850] [Cited by in F6Publishing: 853] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 13. | Moghaddam A, Melum E, Reinton N, Ring-Larsen H, Verbaan H, Bjøro K, Dalgard O. IL28B genetic variation and treatment response in patients with hepatitis C virus genotype 3 infection. Hepatology. 2011;53:746-754. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 14. | Mangia A, Thompson AJ, Santoro R, Piazzolla V, Tillmann HL, Patel K, Shianna KV, Mottola L, Petruzzellis D, Bacca D. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821-87, 827.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 232] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 15. | Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887-1898. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 478] [Cited by in F6Publishing: 463] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 16. | Rao HY, Sun DG, Jiang D, Yang RF, Guo F, Wang JH, Liu F, Zhang HY, Zhang HH, Du SC. IL28B genetic variants and gender are associated with spontaneous clearance of hepatitis C virus infection. J Viral Hepat. 2012;19:173-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 17. | Montes-Cano MA, García-Lozano JR, Abad-Molina C, Romero-Gómez M, Barroso N, Aguilar-Reina J, Núñez-Roldán A, González-Escribano MF. Interleukin-28B genetic variants and hepatitis virus infection by different viral genotypes. Hepatology. 2010;52:33-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 18. | Romero-Gomez M, Eslam M, Ruiz A, Maraver M. Genes and hepatitis C: susceptibility, fibrosis progression and response to treatment. Liver Int. 2011;31:443-460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Barreiro P, Pineda JA, Rallón N, Naggie S, Martín-Carbonero L, Neukam K, Rivero A, Benito JM, Caruz A, Vispo E. Influence of interleukin-28B single-nucleotide polymorphisms on progression to liver cirrhosis in human immunodeficiency virus-hepatitis C virus-coinfected patients receiving antiretroviral therapy. J Infect Dis. 2011;203:1629-1636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 20. | Marabita F, Aghemo A, De Nicola S, Rumi MG, Cheroni C, Scavelli R, Crimi M, Soffredini R, Abrignani S, De Francesco R. Genetic variation in the interleukin-28B gene is not associated with fibrosis progression in patients with chronic hepatitis C and known date of infection. Hepatology. 2011;54:1127-1134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 21. | Falleti E, Bitetto D, Fabris C, Cussigh A, Fornasiere E, Cmet S, Fumolo E, Bignulin S, Fontanini E, Cerutti A. Role of interleukin 28B rs12979860 C/T polymorphism on the histological outcome of chronic hepatitis C: relationship with gender and viral genotype. J Clin Immunol. 2011;31:891-899. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 22. | Asselah T, De Muynck S, Broët P, Masliah-Planchon J, Blanluet M, Bièche I, Lapalus M, Martinot-Peignoux M, Lada O, Estrabaud E. IL28B polymorphism is associated with treatment response in patients with genotype 4 chronic hepatitis C. J Hepatol. 2012;56:527-532. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 23. | Esmat G, El Raziky M, El Kassas M, Hassany M, Gamil ME. The future for the treatment of genotype 4 chronic hepatitis C. Liver Int. 2012;32 Suppl 1:146-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 24. | Kurbanov F, Abdel-Hamid M, Latanich R, Astemborski J, Mohamed M, Mikhail NM, El-Daly M, El-Kafrawy S, Thomas DL, Thio CL. Genetic polymorphism in IL28B is associated with spontaneous clearance of hepatitis C virus genotype 4 infection in an Egyptian cohort. J Infect Dis. 2011;204:1391-1394. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 25. | De Nicola S, Aghemo A, Rumi MG, Galmozzi E, Valenti L, Soffredini R, De Francesco R, Prati GM, D'Ambrosio R, Cheroni C. Interleukin 28B polymorphism predicts pegylated interferon plus ribavirin treatment outcome in chronic hepatitis C genotype 4. Hepatology. 2012;55:336-342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |