Published online Dec 7, 2012. doi: 10.3748/wjg.v18.i45.6651
Revised: August 10, 2012
Accepted: August 25, 2012
Published online: December 7, 2012
AIM: To investigate the clinicopathological features and prognostic value of lysine specific demethylase 1 (LSD1) in hepatocellular carcinoma (HCC).
METHODS: We examined LSD1 expression in 60 paired liver cancer tissues and adjacent noncancerous tissues by quantitative real time polymerase chain reaction (qRT-PCR) and Western blotting. In addition, we analyzed LSD1 expression in 198 HCC samples by immunohistochemistry. The relationship between LSD1 expression, clinicopathological features and patient survival was investigated.
RESULTS: Immunohistochemistry, Western blotting, and qRT-PCR consistently confirmed LSD1 overexpression in HCC tissues compared to adjacent non-neoplastic tissues (P < 0.01). Additionally, immunostaining showed more LSD1-positive cells in the higher tumor stage (T3-4) and tumor grade (G3) than in the lower tumor stage (T1-2, P < 0.001) and tumor grade (G1-2, P < 0.001), respectively. Moreover, HCC patients with high LSD1 expression had significantly lower 5-year overall survival rates (P < 0.001) and lower 5-year disease-free survival rates (P < 0.001), respectively. A Cox proportional hazards model further demonstrated that LSD1 over-expression was an independent predictor of poor prognosis for both 5-year disease-free survival [hazards ratio (HR) = 1.426, 95%CI: 0.672-2.146, P < 0.001] and 5-year overall survival (HR = 2.456, 95%CI: 1.234-3.932, P < 0.001) in HCC.
CONCLUSION: Our data suggest for the first time that the overexpression of LSD1 protein in HCC tissues indicates tumor progression and predicts poor prognosis.
- Citation: Zhao ZK, Yu HF, Wang DR, Dong P, Chen L, Wu WG, Ding WJ, Liu YB. Overexpression of lysine specific demethylase 1 predicts worse prognosis in primary hepatocellular carcinoma patients. World J Gastroenterol 2012; 18(45): 6651-6656
- URL: https://www.wjgnet.com/1007-9327/full/v18/i45/6651.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i45.6651
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide, especially in Asia[1]. In China, HCC ranks behind gastric and esophageal cancer with the third highest mortality rate among all malignant carcinomas, leads to approximately 110 000 deaths every year, and accounts for 45% of total HCC deaths worldwide[2]. Multiple risk factors have been associated with the initiation and development of HCC; these include chronic infection with hepatitis B, C or D virus, aflatoxin, alcohol abuse, hereditary metabolic liver diseases, and diabetes mellitus[1,3]. Like most other cancers, hepatocarcinogenesis is a multistep process that involves multiple genetic alterations that may activate oncogenes and/or inactivate tumor suppressor genes, ultimately leading to the malignant transformation of hepatocytes.
Histone demethylase lysine specific demethylase 1 (LSD1), the first histone demethylase that was discovered as a nuclear homolog of amine oxidases, removes the methyl groups from mono- and dimethylated lysine (Lys) 4 of histone H3 (H3K4me1/2) and Lys9 of histone H3 (H3K9me1/2). LSD1 is essential for mammalian development and is involved in many biological processes, including cell-type differentiation, gene activation and gene repression[4]. A recent study indicated that LSD1 might promote cell phase transition (deficiency in LSD1 led to partial cell cycle arrest in G2/M) and cell proliferation, suggesting that its over-expression might promote tumorigenesis[5]. The expression of LSD1 has been associated with tumor recurrence during therapy in various cancers, further implicating LSD-1 as a tumor promoter[6-8]. A tissue cDNA microarray analysis also demonstrated the presence of LSD1 transactivation in lung and colorectal carcinomas[7]. LSD1 knockdown with small interfering (si) RNAs resulted in the suppression of proliferation of various bladder and lung cancer cell lines[7]. To the best of our knowledge, there is little available data regarding the involvement of LSD1 genes in hepatic tumorigenesis. In this study, we investigated LSD1 expression in HCC and its correlation with the clinicopathological features of patients with HCC, including patient survival.
The study was approved by the Research Ethics Committee of Xinhua Hospital, which is affiliated to Shanghai Jiaotong University School of Medicine, Shanghai, China. Informed consent was obtained from all of the patients. All specimens were handled and made anonymous according to accepted ethical and legal standards.
A total of 198 patients who presented with primary HCC and later underwent curative liver resection at Xinhua Hospital affiliated to Shanghai Jiaotong University School of Medicine, Shanghai, China, were included in this retrospective study. The tissue samples used in this study were retrieved from the tissue bank of the Department of Pathology in the Xinhua Hospital affiliated with Shanghai Jiaotong University School of Medicine. The patients had been diagnosed with HCC between 2001 and 2006. None of the patients recruited in this study had undergone preoperative chemotherapy or radiotherapy. HCC diagnosis was based on World Health Organization criteria. Tumor differentiation was defined according to the Edmondson grading system. Liver function was assessed using the Child-Pugh scoring system. Tumor staging was determined according to the sixth edition of the tumor-node-metastasis (TNM) classification of the International Union against Cancer. The clinicopathological features of 198 patients are summarized in Table 1. In addition, 60 self-pairs of HCC specimens (10 TNM stageI, 16 TNM stage II, 24 TNM stage III, and 10 TNM stage IV) and adjacent non-neoplastic liver tissues were snap frozen in liquid nitrogen and stored at -80 °C following surgery for quantitative real time polymerase chain reaction (qRT-PCR) assay and western blot analysis. The median follow-up period was 8.6 years. Postoperative surveillance included routine clinical and laboratory examinations every third month, computed tomography scans of the abdomen, and radiographs of the chest every third month. After 5 years, the examination interval was extended to 12 mo.
| Characteristics | n | LSD1 (%) | P value | |
| High expression | Low expression | |||
| Gender | ||||
| Male | 101 | 58 | 43 | |
| Female | 97 | 54 | 43 | NS |
| Age (yr) | ||||
| ≥ 50 | 112 | 61 | 51 | |
| < 50 | 86 | 51 | 35 | NS |
| Tumor stage | ||||
| T1 | 39 | 5 | 34 | |
| T2 | 42 | 21 | 21 | |
| T3 | 76 | 47 | 29 | |
| T4 | 41 | 39 | 2 | < 0.001 |
| Tumor grade | ||||
| G1 | 45 | 9 | 34 | |
| G2 | 114 | 63 | 51 | |
| G3 | 39 | 38 | 1 | < 0.001 |
| Growth pattern | ||||
| Trabecular | 147 | 84 | 63 | |
| Nontrabecular | 51 | 28 | 23 | NS |
| Cirrhosis | ||||
| Yes | 151 | 87 | 64 | |
| No | 47 | 25 | 22 | NS |
| Underlying liver disease | ||||
| Alcoholic | 21 | 15 | 6 | |
| Hepatitis B | 136 | 74 | 62 | |
| Hepatitis C | 30 | 17 | 13 | |
| Unknown | 11 | 6 | 5 | NS |
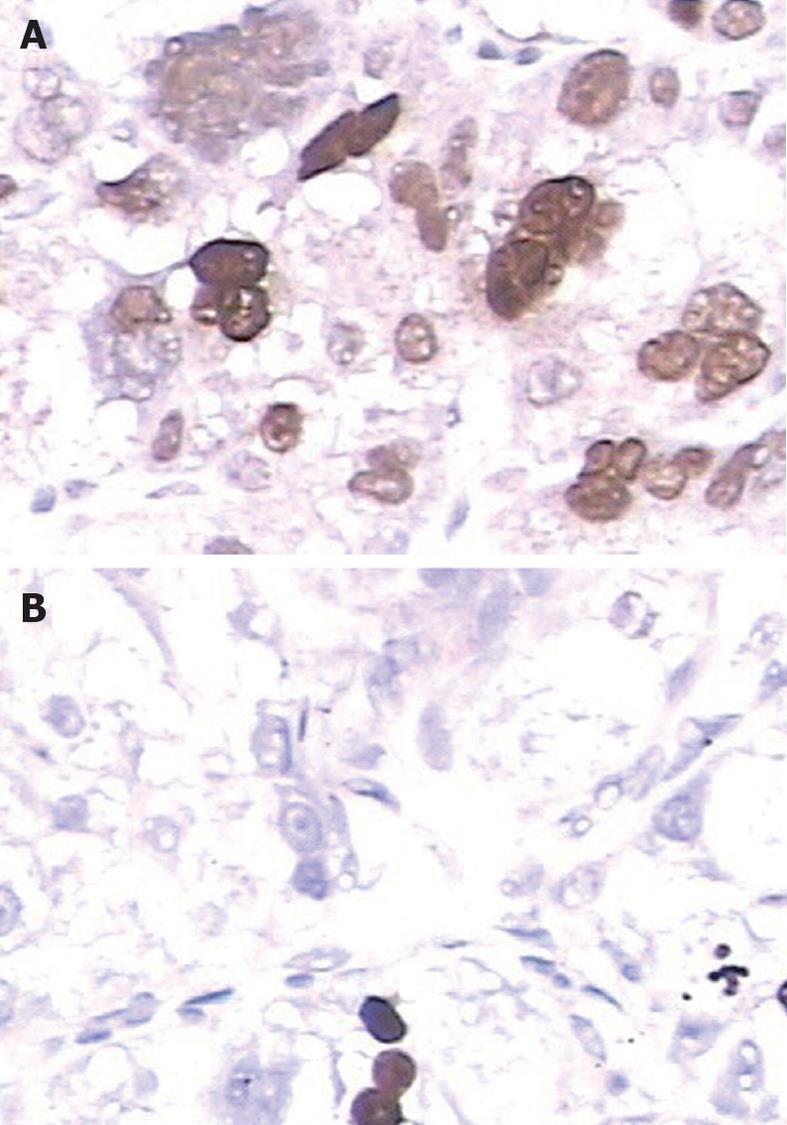
Immunohistochemical staining was carried out following the protocol of our previous study[9-11]. The primary antibody against LSD1 was a rabbit polyclonal antibody (Santa Cruz Biotechnology, Inc., United States) at a dilution of 1:50. The specificity of the primary antibody has been validated by the previous studies of Müller et al[12] and Lü et al[13]. The secondary antibody for the detection of primary antibody was anti-rabbit immunoglobulin G (Sigma, St. Louis, MO, United States). The negative controls were processed in a similar manner with phosphate-buffered saline instead of primary antibody. Further, positive LSD1 expression, as confirmed by western blotting, was used as a positive control for immunostaining. Following hematoxylin counterstaining, immunostaining was scored by two independent experienced pathologists who were blinded to the clinicopathological parameters and clinical outcomes of the patients. The scores of the two pathologists were compared, and any discrepant scores were re-examined for staining by both pathologists until a consensus score was obtained. The number of cells that stained positive for nuclear LSD1 in ten representative microscopic fields was counted, and the percentage of positive cells was calculated. The percentages of cells that were immunoreactive were converted to scores as follows: 0 (0%), 1 (1%-10%), 2 (11%-50%) and 3 (> 50%). Staining intensity was visually scored and stratified as follows: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). A final score was obtained for each case by multiplying the percentage score and the intensity score. Multiplied scores exceeding 5 (median of total scores for LSD1) were considered to indicate low levels of LSD1 expression, while all other scores were considered to indicate high levels of LSD1 expression.
The Western blotting protocol and semiquantitative analysis were carried out following the protocol of Xu et al[14]. LSD1 antibody (rabbit polyclonal antibody, dilution 1:50, Santa Cruz Biotechnology, Inc., United States) was used, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (CW0266, dilution 1:1000, CoWin Biotech) was used as an internal control.
To measure the mRNA expression levels of LSD1, total RNA was extracted from frozen liver tissues using TriZol reagent (Invitrogen) according to the manufacturer’s instructions. The extraction was followed by RT-PCR using the TransStart Green qPCR SuperMix (TransGen Biotech). The primer sequences for LSD1 amplification were 5’-CGAACGCACATCAAGACGA-3’ for the forward primer and 5’-AGGTGAAGGTGGAGTAGAGGC-3’ for the reverse primer. The transcription of GAPDH was used as an internal control for normalization. LSD1 expression levels were calculated relative to GAPDH using the delta-delta computed tomography method[15].
SPSS version 13.0 for Windows (SPSS Inc, IL, United States) and SAS 9.1 (SAS Institute, Cary, NC) were used for statistical analysis. A Fisher’s exact test and χ2 test were performed to assess associations between LSD1 expression and clinicopathological parameters. A Kaplan-Meier method was used for survival analysis, and differences in survival were estimated using the log-rank test. A multivariate survival analysis was performed for all parameters that were significant in the univariate analyses using a Cox regression model. Differences were considered statistically significant when P was less than 0.05.
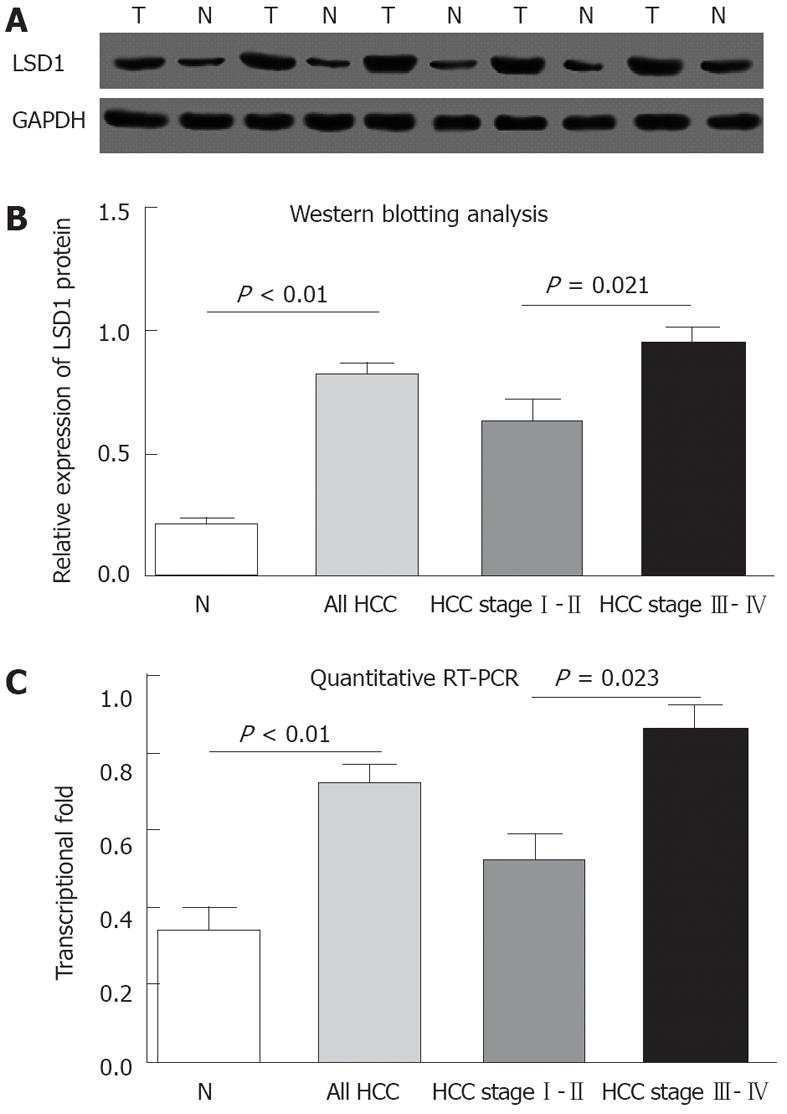
Immunohistochemical analysis revealed that LSD1 staining was mainly localized to the nucleus of noncancerous and malignant epithelial cells (Figure 1). In addition, we found that 112 (56.6%) of 198 HCC tissues had high LSD1 expression, and 86 (43.4%) of 198 HCC tissues had low LSD1 expression. Additionally, 52 (26.3%) of 198 adjacent non-neoplastic liver tissues had high LSD1 expression, and 146 (73.7%) of 198 adjacent nonneoplastic liver tissues had low LSD1 expression. Thus, the LSD1 immunostaining in HCC tissues was significantly higher than the staining in the adjacent non-neoplastic liver tissues (P < 0.01). To confirm LSD1 protein expression by an independent method, Western blotting analysis was performed using 60 self-pairs of HCC and adjacent non-neoplastic liver tissues. Overexpression of LSD1 protein in HCC tissues was compared with adjacent non-neoplastic liver tissues (P < 0.01, Figure 2A and B) using this method, and significantly increased LSD1 mRNA levels were detected by qRT-PCR (P < 0.01, Figure 2C). The expression levels of LSD1 protein and mRNA in high stage (III-IV) HCC tissues were both significantly higher than the levels of LSD1 protein and mRNA in low stage (I-II) HCC tissues (for protein: P = 0.021; for mRNA: P = 0.023; Figure 2B and C).
To evaluate whether LSD1 protein expression was associated with clinicopathological features of patients with HCC, we correlated immunohistochemical LSD1 staining results with T stage, tumor grade, presence of cirrhosis, underlying liver disease, including alcohol abuse, viral hepatitis B and C, sex and age (Table 1). We found more LSD1 positive cells in tissues with higher tumor stages (T3-4) and higher tumor grades (G3) than in the lower tumor stages (T1-2, P < 0.001) and tumor grades (G1-2, P < 0.001), respectively.
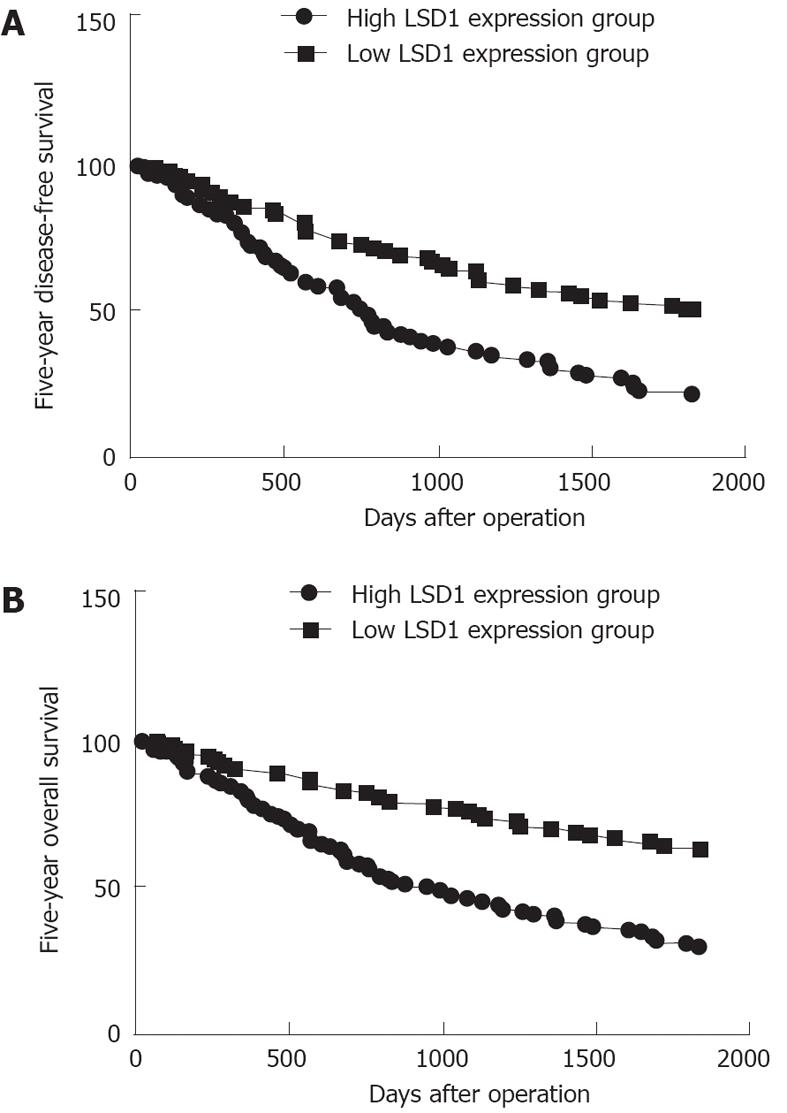
The 5-year disease-free survival in the group with high LSD1 expression was significantly poorer than the disease-free survival in the group with low LSD1 expression (P < 0.001, log-rank test; Figure 3A). A Kaplan-Meier plot of 5-year overall survival curves stratified by LSD1 expression is shown in Figure 3B. There was a significant relationship between LSD1 expression and 5-year overall survival (P < 0.001, log-rank test; Figure 3B). In a multivariate Cox model that included tumor size, tumor stage, tumor grading, presence of cirrhosis, gender, age, and LSD1 staining, we found that LSD1 expression independently indicated poor prognosis for both 5-year disease-free survival [hazards ratio (HR) = 1.426, 95%CI: 0.672-2.146, P < 0.001; Table 2] and 5-year overall survival (HR = 2.456, 95%CI: 1.234-3.932, P < 0.001; Table 2) in patients with HCC.
| Parameter | Five-year overall survival | Five-year disease-free survival | ||||
| P value | HR | 95%CI | P value | HR | 95%CI | |
| Age | 0.775 | 0.867 | 0.463-1.452 | 0.714 | 1.174 | 0.883-1.853 |
| Gender | 0.456 | 1.121 | 0.569-1.867 | 0.634 | 1.126 | 0.684-1.846 |
| Tumor stage | < 0.001 | 1.634 | 1.142-2.537 | < 0.001 | 1.423 | 0.784-2.161 |
| Tumor grade | < 0.001 | 1.154 | 0.647-1.893 | < 0.001 | 1.023 | 0.456-1.638 |
| Presence of cirrhosis | 0.542 | 1.143 | 0.647-1.784 | 0.427 | 1.321 | 0.824-1.917 |
| LSD1 expression | < 0.001 | 2.456 | 1.234-3.932 | < 0.001 | 1.426 | 0.672-2.146 |
Genetic alterations are a hallmark of human cancer. In recent years, the field of cancer genomics has made significant advances in the area of cancer-associated genetic lesion identification. Furthermore, the importance of epigenetic changes that occur during HCC development has also been recognized[16]. Epigenetic changes can take the form of DNA methylation or histone modification[17]. Histone modifications in the form of selective acetylation, phosphorylation, and methylation serve as switches that alter chromatin structure, allowing posttranscriptional activation or repression of downstream proteins[18]. Understanding these epigenetic changes will lead to the identification of novel cancer-related genes that may represent attractive targets for cancer treatment and provide new insights into the biology of hepatic cancers. Thus, an integrative approach to hepatic cancer research that combines epidemiological, genetic and epigenetic information has emerged as an important paradigm for cancer therapy[19]. The methylation status of histone methyltransferases and histone demethylases plays a pivotal role in the regulation of gene expression[20]. LSD1, the first histone demethylase that was discovered as a nuclear homolog of amine oxidases, removes methyl groups from mono- and dimethylated H3K4me1/2 and H3K9me1/2[21].
Epigenetic changes in LSD1 have been shown to play a key role in carcinogenesis[22]. LSD1 can prevent the accumulation of the dimethyl groups of p53, repressing p53-mediated transcriptional up-regulation, preventing apoptosis, and contributing to human carcinogenesis via a chromatin modification mechanism. To date, a few studies have indicated that LSD1 may promote cell phase transition (deficiency in LSD1 led to partial cell cycle arrest in G2/M) and cell proliferation, suggesting that LSD1 over-expression might promote tumorigenesis[5]. The expression of LSD1 has been associated with tumor recurrence during therapy in various cancers, further implicating LSD-1 as a tumor promoter[6-8]. Tissue cDNA microarray analysis also revealed LSD1 transactivation in lung and colorectal carcinomas[7]. Knocking down LSD1 with small interfering RNAs resulted in suppression of proliferation of various bladder and lung cancer cell lines[7]. However, the association between LSD1 and the survival of HCC patients was not well defined. In this study, we investigated the associations between LSD1 expression levels and clinical features of HCC patients.
In order to demonstrate that the epigenetic changes were associated with genetic changes in lung cancer, we first investigated the expression of LSD1 in HCC clinical samples. Previous studies have demonstrated that LSD1 protein and mRNA levels could act as biomarkers for the identification of patients with more aggressive breast cancer, prostate cancer, lung cancer and neuroblastoma[23-26]. In our study, we detected LSD1 by immunohistochemistry analysis, Western blotting, and qRT-PCR. Our results showed that LSD1 immunoreactivity was significantly increased in HCC compared with adjacent non-neoplastic liver tissue in a substantial proportion of cases. The overexpression of LSD1 was observed in tumor tissues with higher tumor stage and higher tumor grade. Additionally, our investigation revealed that high LSD1 expression is associated with a significant trend toward both poorer disease-free survival and poorer overall survival. Our study further confirms that high LSD1 expression independently predicts a higher risk of disease relapse or death after multivariate adjustment for other prognostic factors. These observations support the hypothesis that LSD1 may function as an oncogene in HCC and suggest that LSD1 may play an important role in the tumorigenesis of HCC. However, the role of LSD1 in HCC remains to be elucidated. Our data may offer new insight into LSD1 as a potentially important contributor to the progression of HCC and as a new prognostic factor for HCC. As the 198 cases of the present study were all obtained from the Chinese population, the results reported here should be further confirmed in other populations.
In conclusion, our study suggests that LSD1 is overexpressed in HCC tissues compared with their benign counterparts. To the best of our knowledge, this is the first study evaluating the expression levels of LSD1 mRNA and protein in HCC tissues and the association between these expression levels and clinicopathologic parameters. The most important finding of this study is that LSD1 expression may predict a poorer prognosis for HCC patients after surgery. Further studies are needed to investigate the precise function of LSD1 in the progression of HCC.
Lysine specific demethylase 1 (LSD1) is essential for mammalian development and is involved in many biological processes, such as cell-type differentiation, gene activation and gene repression. Knocking down LSD1 with small interfering RNAs suppressed the proliferation of various bladder and lung cancer cell lines. To be known, little data has been generated with regard to the involvement of LSD1 genes in hepatic tumorigenesis. In this study, the authors investigated LSD1 expression in hepatocellular carcinoma (HCC) and its correlation with clinicopathological features, including the survival of patients with HCC.
The data suggest for the first time that the overexpression of LSD1 protein in HCC tissues may help predict tumor progression and poor prognosis.
The authors examined LSD1 expression in 60 paired liver cancer tissues and adjacent noncancerous tissues by quantitative real time polymerase chain reaction and Western blotting. In addition, authors analyzed LSD1 expression in 198 HCC samples by immunohistochemistry. The relationships between LSD1 expression and both clinicopathological features and patient survival were investigated.
These findings provide evidence that the overexpression of LSD1 serves as a biomarker for poor prognosis in HCC. Thus, authors speculate that LSD1 may be a potential target of anti-angiogenic therapy for HCC.
LSD1, the first histone demethylase that was discovered as a nuclear homolog of amine oxidases, removes the methyl groups from mono- and dimethylated Lysine (Lys) 4 of histone H3 and Lys9 of histone H3.
The authors investigated the expression of LSD1 in HCC and determined its correlation with tumor progression and prognosis. The authors claim that the expression levels of LSD1 protein in HCC tissues correlates with tumor progression and prognosis.
Peer reviewers: Gabriele Grassi, Associate Professor, Department of Medical, Technological and Tran, University Hospital of Cattinara, Strada di Fiume 447, 34100 Trieste, Italy; Vezali Elena, MD, Department of Hepatology, “Hygeia” Diagnostic and Therpaeutic Center of Athens, Eruthrou Staurou 4, 15123 Marousi, Greece
S- Editor Gou SX L- Editor A E- Editor Li JY
| 1. | Schütte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma--epidemiological trends and risk factors. Dig Dis. 2009;27:80-92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 305] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 2. | Li D, Mallory T, Satomura S. AFP-L3: a new generation of tumor marker for hepatocellular carcinoma. Clin Chim Acta. 2001;313:15-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 238] [Cited by in F6Publishing: 227] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 3. | Blum HE, Spangenberg HC. Hepatocellular carcinoma: an update. Arch Iran Med. 2007;10:361-371. [PubMed] [Cited in This Article: ] |
| 4. | Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882-887. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 409] [Cited by in F6Publishing: 425] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 5. | Scoumanne A, Chen X. The lysine-specific demethylase 1 is required for cell proliferation in both p53-dependent and -independent manners. J Biol Chem. 2007;282:15471-15475. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 116] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 6. | Lim S, Janzer A, Becker A, Zimmer A, Schüle R, Buettner R, Kirfel J. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512-520. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 351] [Cited by in F6Publishing: 362] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 7. | Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer. 2011;128:574-586. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 386] [Cited by in F6Publishing: 362] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 8. | Chen PP, Li WJ, Wang Y, Zhao S, Li DY, Feng LY, Shi XL, Koeffler HP, Tong XJ, Xie D. Expression of Cyr61, CTGF, and WISP-1 correlates with clinical features of lung cancer. PLoS One. 2007;2:e534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 116] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 9. | Guo X, Xiong L, Zou L, Zhao J. Upregulation of bone morphogenetic protein 4 is associated with poor prognosis in patients with hepatocellular carcinoma. Pathol Oncol Res. 2012;18:635-640. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 10. | Schmilovitz-Weiss H, Tobar A, Halpern M, Levy I, Shabtai E, Ben-Ari Z. Tissue expression of squamous cellular carcinoma antigen and Ki67 in hepatocellular carcinoma-correlation with prognosis: a historical prospective study. Diagn Pathol. 2011;6:121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 11. | Hong H, Patonay B, Finley J. Unusual reticulin staining pattern in well-differentiated hepatocellular carcinoma. Diagn Pathol. 2011;6:15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 12. | Müller P, Crofts JD, Newman BS, Bridgewater LC, Lin CY, Gustafsson JA, Ström A. SOX9 mediates the retinoic acid-induced HES-1 gene expression in human breast cancer cells. Breast Cancer Res Treat. 2010;120:317-326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 13. | Lü B, Fang Y, Xu J, Wang L, Xu F, Xu E, Huang Q, Lai M. Analysis of SOX9 expression in colorectal cancer. Am J Clin Pathol. 2008;130:897-904. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 14. | Xu MZ, Yao TJ, Lee NP, Ng IO, Chan YT, Zender L, Lowe SW, Poon RT, Luk JM. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009;115:4576-4585. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 364] [Cited by in F6Publishing: 401] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 15. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117419] [Cited by in F6Publishing: 121468] [Article Influence: 5281.2] [Reference Citation Analysis (0)] |
| 16. | Piperi C, Vlastos F, Farmaki E, Martinet N, Papavassiliou AG. Epigenetic effects of lung cancer predisposing factors impact on clinical diagnosis and prognosis. J Cell Mol Med. 2008;12:1495-1501. [PubMed] [Cited in This Article: ] |
| 17. | Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, Wang Q, Chia D, Goodglick L, Kurdistani SK. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174:1619-1628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 358] [Cited by in F6Publishing: 382] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 18. | Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829-833. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 437] [Cited by in F6Publishing: 428] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 19. | Risch A, Plass C. Lung cancer epigenetics and genetics. Int J Cancer. 2008;123:1-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 169] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 20. | Ueda R, Suzuki T, Mino K, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. Identification of cell-active lysine specific demethylase 1-selective inhibitors. J Am Chem Soc. 2009;131:17536-17537. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 157] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 21. | Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941-953. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2977] [Cited by in F6Publishing: 3032] [Article Influence: 159.6] [Reference Citation Analysis (0)] |
| 22. | Lim S, Metzger E, Schüle R, Kirfel J, Buettner R. Epigenetic regulation of cancer growth by histone demethylases. Int J Cancer. 2010;127:1991-1998. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 23. | Zhu Q, Huang Y, Marton LJ, Woster PM, Davidson NE, Casero RA. Polyamine analogs modulate gene expression by inhibiting lysine-specific demethylase 1 (LSD1) and altering chromatin structure in human breast cancer cells. Amino Acids. 2012;42:887-898. [PubMed] [Cited in This Article: ] |
| 24. | Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, Ora I, Pajtler K, Klein-Hitpass L, Kuhfittig-Kulle S. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69:2065-2071. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 329] [Cited by in F6Publishing: 335] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 25. | Suikki HE, Kujala PM, Tammela TL, van Weerden WM, Vessella RL, Visakorpi T. Genetic alterations and changes in expression of histone demethylases in prostate cancer. Prostate. 2010;70:889-898. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 26. | Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X, Song Y. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS One. 2012;7:e35065. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 217] [Cited by in F6Publishing: 215] [Article Influence: 17.9] [Reference Citation Analysis (0)] |