Published online Jun 14, 2012. doi: 10.3748/wjg.v18.i22.2850
Revised: September 13, 2011
Accepted: March 10, 2012
Published online: June 14, 2012
AIM: To investigate whether DNA-dependent activator of interferon-regulatory factors (DAI) inhibits hepatitis B virus (HBV) replication and what the mechanism is.
METHODS: After the human hepatoma cell line Huh7 was cotransfected with DAI and HBV expressing plasmid, viral protein (HBV surface antigen and HBV e antigen) secretion was detected by enzyme-linked immunosorbent assay, and HBV RNA was analyzed by real-time polymerase chain reaction and Northern blotting, and viral DNA replicative intermediates were examined by Southern blotting. Interferon regulatory factor 3 (IRF3) phosphorylation and nuclear translocation were analyzed via Western blotting and immunofluorescence staining respectively. Nuclear factor-κB (NF-κB) activity induced by DAI was detected by immunofluorescence staining of P65 and dual luciferase reporter assay. Transwell co-culture experiment was performed in order to investigate whether the antiviral effects of DAI were dependent on the secreted cytokines.
RESULTS: Viral protein secretion was significantly reduced by 57% (P < 0.05), and the level of total HBV RNA was reduced by 67% (P < 0.05). The viral core particle-associated DNA was also dramatically down-regulated in DAI-expressing Huh7 cells. Analysis of involved signaling pathways revealed that activation of NF-κB signaling was essential for DAI to elicit antiviral response in Huh7 cells. When the NF-κB signaling pathway was blocked by a NF-κB signaling suppressor (IκBα-SR), the anti-HBV activity of DAI was remarkably abrogated. The inhibitory effect of DAI was independent of IRF3 signaling and secreted cytokines.
CONCLUSION: This study demonstrates that DAI can inhibit HBV replication and the inhibitory effect is associated with activation of NF-κB but independent of IRF3 and secreted cytokines.
- Citation: Chen QY, Liu YH, Li JH, Wang ZK, Liu JX, Yuan ZH. DNA-dependent activator of interferon-regulatory factors inhibits hepatitis B virus replication. World J Gastroenterol 2012; 18(22): 2850-2858
- URL: https://www.wjgnet.com/1007-9327/full/v18/i22/2850.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i22.2850
Hepatitis B virus (HBV) is a noncytopathic DNA virus which belongs to the Hepadnaviridae family. Infection of HBV results in acute or chronic hepatitis, liver failure, and hepatocellular carcinoma[1-2]. HBV clearance is usually associated with a multispecific CD4+ and CD8+ T-cell response coordinated with an effective humoral immune component[3-5]. However, a growing body of evidence suggests that the innate immune response is important for limiting viral replication. Expression of key proteins in pattern recognition system, such as RNA sensor melanoma differentiation-associated gene-5, the caspase recruitment domain of retinoic acid inducible gene I and the adaptor protein, myeloid differentiation primary response protein 88 (MyD88), and interferon-β promoter stimulator 1 (IPS-1) can activate innate immune response and inhibit HBV replication in human hepatocyte-derived cells[6,7].
DNA-dependent activator of interferon-regulatory factor (DAI/DLM-1/ZBP1) is the first identified sensor of cytosolic dsDNA. Recent studies have demonstrated that DAI can initiate innate immune responses, including the induction of type I interferon (IFN) genes, independently of Toll-like receptor 9[8-10]. It was reported that herpes simplex virus 1 production was notably higher in DAI blocked L929 cells[10]. As DAI is highly expressed in the differentiated hepatocyte after interferon treatment[11], it is hypothesized that DAI could be a protein possessing antiviral activity against HBV in human hepatocytes.
The aim of the present study was to determine whether DAI can inhibit HBV replication and what the underlying molecular mechanism is. We found that expression of DAI could inhibit HBV gene expression and replication noncytopathically in Huh7 cells. Further study revealed that activation of nuclear factor-κB (NF-κB) signaling was essential for DAI to elicit antiviral responses, but this inhibitory effect is independent of cytokines’ secretion.
Huh7 and HEK 293T cells were obtained from American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium (Gibco). For plasmid transfections, we used Fugene reagent (Roche), according to the manufacturer’s protocol. The transfection efficiency was normalized by detecting the expression of green fluorescent protein (GFP) after transfection of equal amount of plasmid GFP.
Transwell plate (Cat. No. 3450) was purchased from Corning Company (New York, the United States). The transwell chamber contains 0.4 μm pore polyester membrane which is optimal for cell attachment and growth and is permeable for flow of liquids. The cytokines can transfer through the membrane freely while the cells are blocked.
pCAGGS-hemagglutinin (HA)-DAI encoding the whole transcript of DAI was kindly provided by Professor Tadatsugu Taniguchi[9]. pHBV1.3 containing a 1.3-copy of the HBV genome was described previously. The plasmid pCMV-IκBα-SR expresses a repressor form of IκBα in which serines 32 and 36 were mutated to alanine[12]. The NF-κB-dependent luciferase reporter plasmid pNF-κB-Luc was obtained from Stratagene Corporation (La Jolla, CA, the United States). pIRF-3 and pIRF-3ΔN were provided by John Hiscott[13]. The cytokines IFN-α and tumor growth factor (TGF)-α were purchased from R and D Company (Lorton, VA, the United States).
Total RNA was extracted directly using TRIzol reagent (Invitrogen), and reversely transcribed to cDNA using a complementary DNA (cDNA) synthesis kit (Fermentas) according to the manufacturer’s instructions. The cDNA was mixed with SYBR Green polymerase chain reaction (PCR) Master Mix (Toyoba) and subjected to real-time PCR using the ABI PRISM 7500 (Applied Biosystems). Cellular glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA from the same cDNA was used as an internal control. The primers specific for HBV and GAPDH are available upon request. Forty cycles of PCR were performed with cycling conditions of 15 s at 95 °C, 20 s at 55 °C, 25 s at 72 °C,and 35 s at 79 °C to detect signal.
HBV DNA replicative intermediates in HBV core particles isolated from transfected Huh7 cells were analyzed using Southern blotting. The intracellular viral DNA was extracted as previously reported[14].
The normalized viral DNA replicative intermediates were electrophoresed onto 1% agarose gel. Then DNA was blotted onto a positive nylon membrane (Roche) in 20 × SSC. After fixing at 120 °C for 30 min, the membrane was prehybridized for 1 h at 42 °C in ULTRAhyb hybridization solution, and then hybridized with full-length HBV DNA probes labeled with (α-32P) deoxycytidine triphosphate (dCTP) by hexamer random labeling kit (Roche) under the same condition of prehybridization at 42 °C for 16 h. After stringent washing at 68 °C, signals were detected by autoradiography.
Total RNA was extracted directly from transfected cells using TRIzol reagent. Ten μg total cytoplasmic RNA was electrophoresed on 1% formaldehyde-agarose gel and then transferred to nylon membranes (Roche). Hybridization was undertaken as described above using (α-32P) dCTP-labeled full-length HBV DNA probes. To normalize the total quantity of RNA loaded on each gel, blots were stripped and rehybridized with (α-32P) dCTP-labeled GAPDH probes.
Cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted electrophoretically onto a nitrocellulose membrane (Whatman). The membrane was blocked with phosphate-buffered saline (PBS) containing 5% skim milk and then incubated overnight with 1:1000 anti-HA (Roche), 1:2000 anti-GFP (Sigma), 1:1000 anti-phospho-IRF3 (Cell signaling), 1:1000 anti-IRF3 (Santa Cruz), 1:10000 anti-β actin antibody (Sigma), then washed three times in PBST (0.05% Tween 20 in PBS), and incubated with peroxidase conjugated secondary antibody (1:2000) for 1 h. After further washing with PBST, chemiluminescence detection was carried out using enhanced chemiluminescence detection reagents.
The HBV surface antigen (HBsAg) and HBV e antigen (HBeAg) levels in the supernatants obtained from HBV transfected Huh7 cells were detected using a standard enzyme-linked immunosorbent assay (ELISA) (Sino-American Biotech). The assay was performed according to the manufacture’s protocol. All experiments were performed at least three times.
Huh7 cells were fixed with 3.5% paraformaldehyde for 10 min. The cells were permeabilized with 0.1% Triton X-100 for 5 min at room temperature and incubated in blocking buffer supplemented with 3% bovine serum albumin (Sigma) for 1 h at room temperature. Interferon regulatory factor 3 (IRF3) was detected by staining with rabbit anti-human IRF3 (1:200 dilution, Santa Cruz), followed by Cy3-coupled goat anti-rabbit IgG (1:500 dilution, Jackson Immunologicals). Flag-tagged IPS1 was detected by staining with mouse anti-Flag antibody (1:2000, Sigma), followed by Alexa488-coupled goat anti-mouse IgG (1:200, Jackson Immunologicals). P65 was detected by staining with diluted (1:100) rabbit anti-human P65 (cell signaling) and Cy3-coupled goat anti-rabbit IgG (1:500). The nuclei were counterstained with 10 μg/mL 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma). After incubation with the secondary antibodies, the cells were visualized under a confocal laser scanning microscope.
To measure report gene activation, 293T cells seeded into 24-well plates at density of 1 × 105 cells/well were transiently transfected with NF-κB dependent luciferase reporter plasmid 100 ng pNF-κB-Luc, 10 ng renilla luciferase-HSV thymidine kinase promoter (expressing Renilla luciferase, Promega) together with pCAGGS-HA-DAI or the empty vector pCAGGS-HA at the indicated amount. The cells were lysed and analyzed for firefly luciferase and renilla luciferase activity (Promega). The results were reported as the normalized mean ± SD.
Results were reported as means ± SD. T tests were applied for comparisons between groups; and P < 0.05 was considered statistically significant.
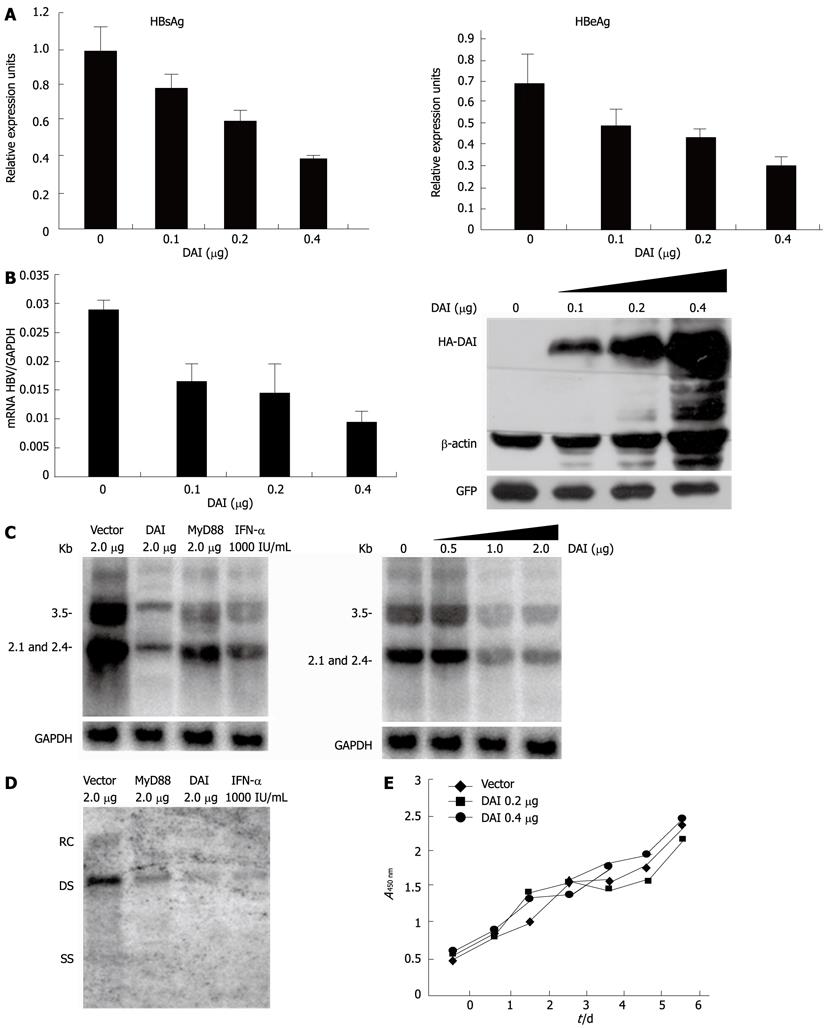
To investigate the antiviral activity of DAI against HBV, we firstly examined the effect of DAI on the synthesis of HBV proteins. HBV-replicating plasmid HBV1.3 was co-transfected with either empty vector or HA-DAI into Huh7 cells. Supernatants were collected and HBsAg and HBeAg were analyzed by standard ELISA immunoassay. Compared with the control, the secretion of HBsAg was reduced by 17%, 33% and 57% and secretion of HBeAg was reduced by 25%, 34% and 57% when the increasing amount of DAI was transfected (Figure 1A). In order to study the inhibitory effect of DAI on HBV RNA transcription, the HBV RNA level was examined by quantitative real-time PCR. Results showed that HBV RNA level was also decreased by 44%, 51%, and 67% with an increased level of DAI expression. Expression of DAI in Huh7 cells was monitored by Western blotting (Figure 1B). To further investigate the effect of DAI on HBV viral RNA transcription, Northern blotting analysis was employed. As MyD88 has been reported as interferon inducible protein which can inhibit HBV replication[6,7], MyD88 and 1000 IU/mL IFN-α treatment were included as positive controls. As shown in Figure 1C, expression of DAI dramatically reduced HBV RNA level. To investigate the influence of DAI on HBV replication, Southern blotting was performed to analyze the viral DNA replicative intermediates which were extracted from core particles. As shown in Figure 1D, the HBV core particle-associated DNA was significantly reduced. These results suggested that viral genome replication, viral RNA transcription and viral protein expression were all downregulated by DAI.
To exclude the possibility that the reduction of HBV RNA and DNA in Huh7 cells was due to cell death induced by DAI, the growth of DAI-expressing Huh7 cells was examined by cell counting assay for 6 d. Results demonstrated that DAI did not obviously affect cell growth (Figure 1E). Taken together, DAI can inhibit HBV gene expression and replication noncytopathically in Huh7 cells.
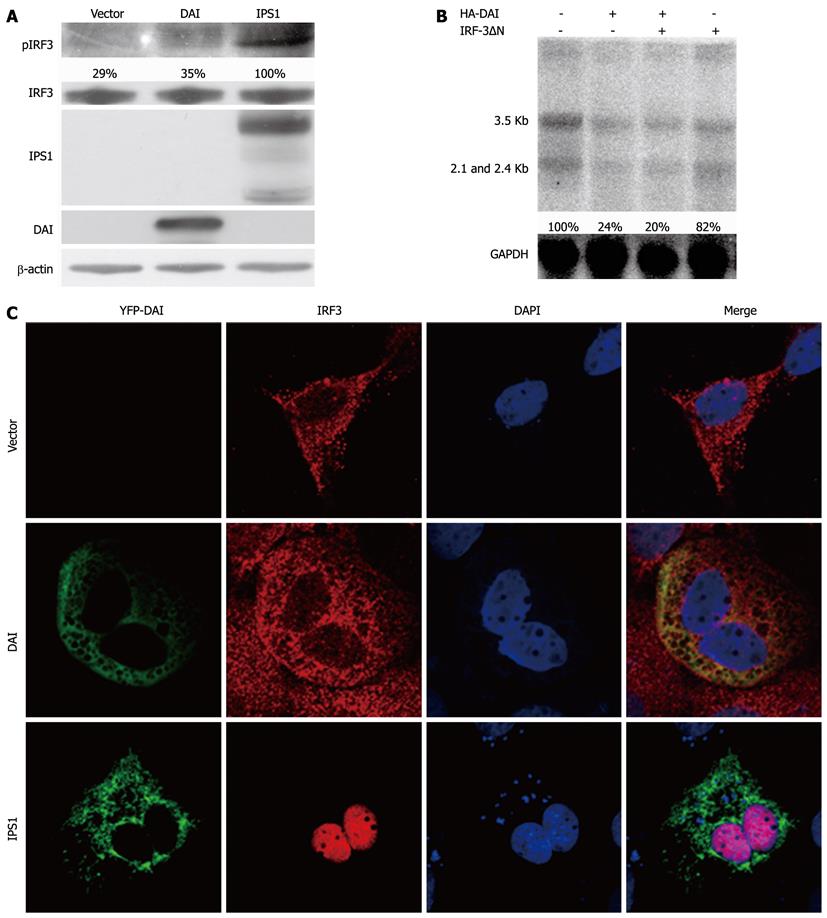
The activation of innate immune system by DAI was through IRF3 or NF-κB mediated signaling pathways[10]. To investigate the possible effect of the pathways DAI on it, we firstly examined the activation of IRF3 after overexpression of DAI. IPS1, which can activate IRF-3 signaling pathway, was set as positive control[15]. The results showed that DAI cannot induce the phosphorylation of IRF-3 (Figure 2A). Furthermore, as shown in Figure 2C, nuclear translocation of IRF-3 was not observed after DAI expression. These results indicated that DAI cannot activate IRF-3. To further confirm that DAI-mediated inhibition of HBV replication is not associated with IRF-3, an IRF-3 dominant negative plasmid (IRF-3ΔN), in which the DNA binding domain was removed to express the repressor form of IRF-3, was used[13]. Results suggested that when the IRF-3 pathway was blocked by IRF-3ΔN, the inhibitory effect of DAI on HBV replication was not affected (Figure 2B). Taken together, inhibition of HBV replication by DAI was not associated with activation of IRF3 pathway.
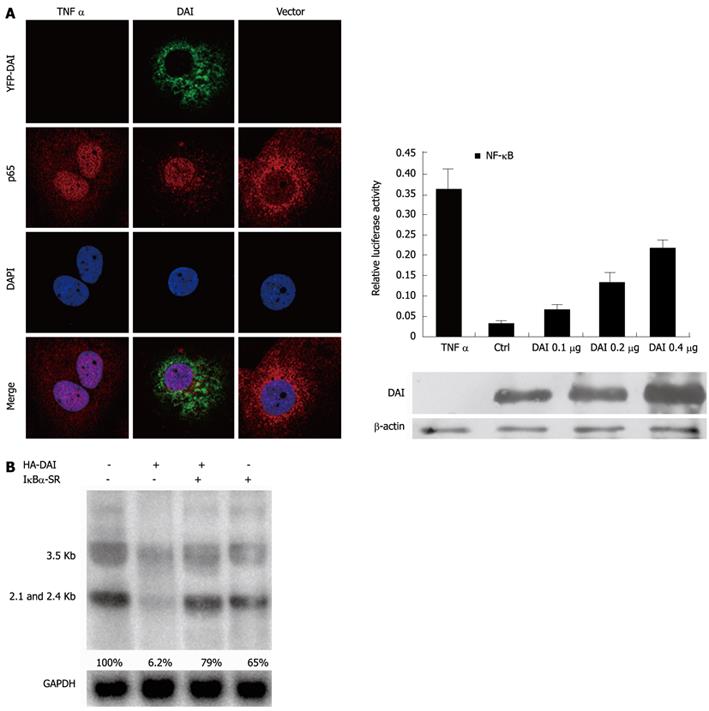
In addition to IRF-3 signaling pathway, NF-κB is another key pathway induced by DAI to activate antiviral innate immunity. In most cells, NF-κB signaling pathway complexes are inactive, residing primarily in the cytoplasm in a complex with the inhibitory IκB proteins. Once activated, NF-κB is released from the IκBα and translocated to nucleus, where it binds to specific κB sequences in the promoter or enhancer regions to induce the expression of multiple target genes[14,16]. To investigate if NF-κB was activated by DAI, we firstly examined the translocation of NF-κB p65. Yellow fluorescent protein-DAI and control DNA was transfected into Huh7 cells and TNF-α treatment was included as positive control. As expected, nuclear translocation of p65 was observed in both DAI-expressing and TNF-α-treated cells. Furthermore, a NF-κB-dependent luciferase reporter plasmid (pNF-κB-Luc) was used to detect NF-κB activity after overexpression of DAI. Results showed that DAI increased the NF-κB-dependent luciferase activity in a dose dependent manner (Figure 3A), suggesting that DAI can induce the activation of NF-κB signaling pathway.
To further confirm that suppression of HBV replication by DAI was NF-κB-dependent, we used a NF-κB signaling suppressor IκBα-SR, in which Ser32 and Ser36 residues critical for phosphorylation are replaced by alanine[12]. As shown in Figure 3B, the inhibition of HBV RNA by DAI was reversed in the presence of IκBα-SR.
In conclusion,these data demonstrated that the activation of NF-κB signaling pathway played an indispensable role in DAI-mediated suppression of HBV replication.
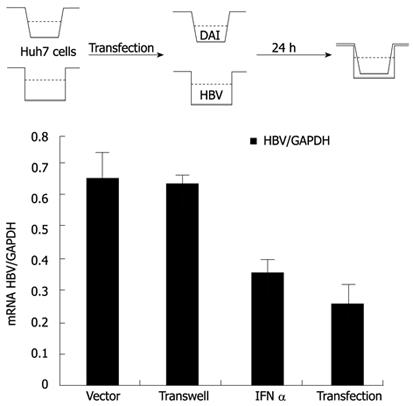
After NF-κB was activated, IFNs, chemokines and other pro-inflammatory cytokines could be induced, which might be directly involved in inhibiting viral infection[17]. To investigate whether the observed antiviral effects of DAI were dependent on the secreted cytokines, transwell co-culture experiments were conducted. The IFN-α treatment was set as positive control. We found that the obvious inhibitory effects of DAI on HBV replication could be observed in the HBV and HA-DAI directly co-transfected cells, but not in transwell co-cultured Huh7 cells (Figure 4), indicating a secreted cytokine-independent mechanism of inhibiting HBV replication by DAI.
The above results suggested that the observed inhibition of HBV replication by DAI was most likely due to some inducible intracellular factor(s), rather than secreted cytokines.
After viral infection, innate immune receptors can detect the invading virus and subsequently initiate the synthesis of IFN and protective cellular genes to directly limit viral replication[18,19]. DAI is the first identified sensor of cytosolic dsDNA, which elicits innate immune responses and induces type I IFN to control viral replication. In this study, we found that DAI could inhibit HBV replication in Huh7 cells, and further study revealed that NF-κB signaling pathway was essential for this inhibition.
NF-κB signaling pathway plays pivotal roles in mediating inflammation, immune responses to pathogen infections, proliferation, apoptosis, and other cellular activities. The activation of NF-κB presents different results for different viruses. Some viruses activate NF-κB pathway to improve their transcription and replication[20]. Under other conditions, activation of NF-κB can repress viral replication. For example, NF-κB activation can mediate inhibition of human cytomegalovirus replication[21]. Rotavirus could antagonize cellular antiviral responses by inhibiting the nuclear accumulation of NF-κB[22]. As far as HBV is concerned, on one hand, viral replication itself can activate NF-κB, and on the other hand, upregulation of NF-κB by some host cytokines’ stimulation has shown to be an inhibitory factor. For example, TNF-α could inhibit HBV replication by activating NF-κB signaling[23]. Besides, activation of NF-κB is also required for MyD88, IPS-1 and TRIF to elicit antiviral response to limit HBV replication[23]. In this study, we also found that DAI inhibited HBV replication via activating NF-κB signaling. Therefore, we speculated that the NF-κB signaling might be a common pathway for host to inhibit the replication of HBV and other DNA viruses.
The activation of NF-κB is associated with increased transcription of genes encoding chemokines, such as IL-8, MCP-1, cytokines such as IL-6, TNF-α, IFNs, adhesion molecules (intercellular adhesion molecule 1 and vascular cell adhesion molecule-1), enzymes that produce secondary inflammatory mediators and inhibitors of apoptosis[24-26]. These molecules are important components of the innate immune response to invading microorganisms. When further exploring the mechanisms responsible for suppression of viral replication after NF-κB activation, we speculated that cytokines, especially type I interferon, may be the direct effector to inhibit HBV replication. However, we did not detect the induction of IFN in DAI-expressing cells. By transwell experiment, we found that the secreted cytokines were not required for the inhibition of HBV replication by DAI (Figure 4). Interestingly, some components of pattern-recognition receptor system, such as MyD88, IPS-1 and TRIF,could also control HBV replication in a cytokine independent manner[24]. It is possible that there is a common strategy to inhibit HBV by these functional proteins. In addition, the antiviral factor downstream of NF-κB induced by DAI is worthy to be further explored.
In summary, this study demonstrates that DAI is a cellular antiviral protein. When expressed in Huh7 cells, DAI activated NF-κB but not IRF-3 signaling to suppress HBV replication. This inhibitory effect is independent of secreted cytokines. The findings could potentially lead to the development of novel therapies that induce the host cytoplasmic antiviral protein to control HBV infections.
The hepatitis B virus (HBV) is a DNA virus that replicates its genome via an RNA intermediate using reverse transcription. Chronic infection with this virus can result in cirrhosis and hepatocellular carcinoma. Nowadays, more and more evidence suggests that the innate immune response is important for limiting viral replication. Pattern recognition receptors play a pivotal role in host innate immune responses against microbial infection. DNA-dependent activator of interferon-regulatory factor (DAI/DLM-1/ZBP1) is a potent activator of immune responses during infection or tissue damage.
Expression of key proteins in pattern recognition system, such as RNA sensor melanoma differentiation-associated gene-5, the caspase recruitment domain of retinoic acid inducible gene I and the adaptor protein, and myeloid differentiation primary response protein88 can activate innate immune response and inhibit HBV replication in human hepatocyte-derived cells. DAI is the first identified sensor of cytosolic dsDNA. Recent studies have demonstrated that DAI can initiate innate immune responses, including the induction of type I interferon genes, independently of Toll-like receptor 9. The authors hypothesize that DAI could be a protein possessing antiviral activity against HBV replication.
The aim of the present study was to determine whether DAI can inhibit HBV replication and what the underlying molecular mechanism is. The authors found that expression of DAI could inhibit HBV gene expression and replication noncytopathically in Huh7 cells. Further study revealed that activation of Nuclear factor-κB signaling was essential for DAI to elicit antiviral responses, but this inhibitory effect was independent of cytokines’ secretion.
The study could potentially lead to the development of novel therapies that induce the host cytoplasmic antiviral protein to control HBV infections.
The study describing the inhibitory role of DAI on HBV replication is generally well-performed and the results support the conclusions reached. The manuscript is well-written.
Peer reviewer: Ourania M Andrisani, PhD, Professor, B038 Hansen Bldg, Center for Cancer Research, Purdue University, West Lafayette, IN 47907, United States
S- Editor Shi ZF L- Editor Ma JY E- Editor Zhang DN
| 1. | Dienstag JL. Hepatitis B virus infection. N Engl J Med. 2008;359:1486-1500. [PubMed] [Cited in This Article: ] |
| 2. | Malik R, Kennedy P, Suri D, Brown A, Goldin R, Main J, Thomas H, Thursz M. The role of liver fibrosis assessment in the management of patients with chronic hepatitis B infection: lessons learned from a single centre experience. Hepat Res Treat. 2011;2011:524027. [PubMed] [Cited in This Article: ] |
| 3. | Ferrari C, Penna A, Bertoletti A, Valli A, Antoni AD, Giuberti T, Cavalli A, Petit MA, Fiaccadori F. Cellular immune response to hepatitis B virus-encoded antigens in acute and chronic hepatitis B virus infection. J Immunol. 1990;145:3442-3449. [PubMed] [Cited in This Article: ] |
| 4. | Rehermann B, Fowler P, Sidney J, Person J, Redeker A, Brown M, Moss B, Sette A, Chisari FV. The cytotoxic T lymphocyte response to multiple hepatitis B virus polymerase epitopes during and after acute viral hepatitis. J Exp Med. 1995;181:1047-1058. [PubMed] [Cited in This Article: ] |
| 5. | Thimme R, Wieland S, Steiger C, Ghrayeb J, Reimann KA, Purcell RH, Chisari FV. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol. 2003;77:68-76. [PubMed] [Cited in This Article: ] |
| 6. | Lin S, Wu M, Xu Y, Xiong W, Yi Z, Zhang X, Zhenghong Y. Inhibition of hepatitis B virus replication by MyD88 is mediated by nuclear factor-kappaB activation. Biochim Biophys Acta. 2007;1772:1150-1157. [PubMed] [Cited in This Article: ] |
| 7. | Guo H, Jiang D, Ma D, Chang J, Dougherty AM, Cuconati A, Block TM, Guo JT. Activation of pattern recognition receptor-mediated innate immunity inhibits the replication of hepatitis B virus in human hepatocyte-derived cells. J Virol. 2009;83:847-858. [PubMed] [Cited in This Article: ] |
| 8. | Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci USA. 2008;105:5477-5482. [PubMed] [Cited in This Article: ] |
| 9. | Takaoka A, Taniguchi T. Cytosolic DNA recognition for triggering innate immune responses. Adv Drug Deliv Rev. 2008;60:847-857. [PubMed] [Cited in This Article: ] |
| 10. | Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501-505. [PubMed] [Cited in This Article: ] |
| 11. | Wieland SF, Vega RG, Müller R, Evans CF, Hilbush B, Guidotti LG, Sutcliffe JG, Schultz PG, Chisari FV. Searching for interferon-induced genes that inhibit hepatitis B virus replication in transgenic mouse hepatocytes. J Virol. 2003;77:1227-1236. [PubMed] [Cited in This Article: ] |
| 12. | Traenckner EB, Pahl HL, Henkel T, Schmidt KN, Wilk S, Baeuerle PA. Phosphorylation of human I kappa B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis and NF-kappa B activation in response to diverse stimuli. EMBO J. 1995;14:2876-2883. [PubMed] [Cited in This Article: ] |
| 13. | Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986-2996. [PubMed] [Cited in This Article: ] |
| 14. | Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280-288. [PubMed] [Cited in This Article: ] |
| 15. | Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981-988. [PubMed] [Cited in This Article: ] |
| 16. | Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195-2224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3002] [Cited by in F6Publishing: 3069] [Article Influence: 153.5] [Reference Citation Analysis (0)] |
| 17. | Sen GC, Sarkar SN. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr Top Microbiol Immunol. 2007;316:233-250. [PubMed] [Cited in This Article: ] |
| 18. | Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370-383. [PubMed] [Cited in This Article: ] |
| 19. | Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Curr Opin Immunol. 2008;20:17-22. [PubMed] [Cited in This Article: ] |
| 20. | Hayashi T, Nishitsuji H, Takamori A, Hasegawa A, Masuda T, Kannagi M. DNA-dependent activator of IFN-regulatory factors enhances the transcription of HIV-1 through NF-κB. Microbes Infect. 2010;12:937-947. [PubMed] [Cited in This Article: ] |
| 21. | Eickhoff JE, Cotten M. NF-kappaB activation can mediate inhibition of human cytomegalovirus replication. J Gen Virol. 2005;86:285-295. [PubMed] [Cited in This Article: ] |
| 22. | Holloway G, Truong TT, Coulson BS. Rotavirus antagonizes cellular antiviral responses by inhibiting the nuclear accumulation of STAT1, STAT2, and NF-kappaB. J Virol. 2009;83:4942-4951. [PubMed] [Cited in This Article: ] |
| 23. | Biermer M, Puro R, Schneider RJ. Tumor necrosis factor alpha inhibition of hepatitis B virus replication involves disruption of capsid Integrity through activation of NF-kappaB. J Virol. 2003;77:4033-4042. [PubMed] [Cited in This Article: ] |
| 24. | Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225-260. [PubMed] [Cited in This Article: ] |
| 25. | Melotti P, Nicolis E, Tamanini A, Rolfini R, Pavirani A, Cabrini G. Activation of NF-kB mediates ICAM-1 induction in respiratory cells exposed to an adenovirus-derived vector. Gene Ther. 2001;8:1436-1442. [PubMed] [Cited in This Article: ] |
| 26. | Zerfaoui M, Suzuki Y, Naura AS, Hans CP, Nichols C, Boulares AH. Nuclear translocation of p65 NF-kappaB is sufficient for VCAM-1, but not ICAM-1, expression in TNF-stimulated smooth muscle cells: Differential requirement for PARP-1 expression and interaction. Cell Signal. 2008;20:186-194. [PubMed] [Cited in This Article: ] |