Published online Jul 28, 2010. doi: 10.3748/wjg.v16.i28.3553
Revised: February 3, 2010
Accepted: February 10, 2010
Published online: July 28, 2010
AIM: To investigate aberrant DNA methylation of CpG islands and subsequent low- or high-level DNA microsatellite instability (MSI) which is assumed to drive colon carcinogenesis.
METHODS: DNA of healthy individuals, adenoma (tubular or villous/tubulovillous) patients, and colorectal carcinoma patients who underwent colonoscopy was used for assessing the prevalence of aberrant DNA methylation of human DNA mismatch repair gene mutator L homologue 1 (hMLH1), Cyclin-dependent kinase inhibitor 2A (CDKN2A/p16), and O-6-methylguanine DNA methyltransferase (MGMT), as well as their relation to MSI.
RESULTS: The frequency of promoter methylation for each locus increased in the sequence healthy tissue/adenoma/carcinoma. MGMT showed the highest frequency in each group. MGMT and CDKN2A/p16 presented a statistically significant increase in promoter methylation between the less and more tumorigenic forms of colorectal adenomas (tubular vs tubullovillous and villous adenomas). All patients with tubulovillous/villous adenomas, as well as all colorectal cancer patients, showed promoter methylation in at least one of the examined loci. These findings suggest a potentially crucial role for methylation in the polyp/adenoma to cancer progression in colorectal carcinogenesis. MSI and methylation seem to be interdependent, as simultaneous hMLH1, CDKN2A/p16, and MGMT promoter methylation was present in 8/9 colorectal cancer patients showing the MSI phenotype.
CONCLUSION: Methylation analysis of hMLH1, CDKN2A/p16, and MGMT revealed specific methylation profiles for tubular adenomas, tubulovillous/villous adenomas, and colorectal cancers, supporting the use of these alterations in assessment of colorectal tumorigenesis.
-
Citation: Psofaki V, Kalogera C, Tzambouras N, Stephanou D, Tsianos E, Seferiadis K, Kolios G. Promoter methylation status of
hMLH1 ,MGMT , andCDKN2A /p16 in colorectal adenomas. World J Gastroenterol 2010; 16(28): 3553-3560 - URL: https://www.wjgnet.com/1007-9327/full/v16/i28/3553.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i28.3553
Chromosomal instability and microsatellite instability (MSI) are two well-known pathways of colorectal carcinogenesis[1,2]. Recently, the CpG island methylation phenotype has been added to these two pathways. This novel pathway is characterized by the concordant methylation of the promoter regions of multiple genes that play a role in carcinogenesis[3], an alteration that has also been associated with the process of aging. CpG island DNA methylation, the most extensively studied epigenetic alteration in neoplasms, represses gene transcription by excessive and aberrant methylation of CpG-rich regions, called “CpG islands”, in the 5’ region of genes, leading to transcriptional silencing of the promoter and therefore inactivation of the gene[4,5].
Aberrant DNA methylation of CpG islands (CpG island methylator phenotype, CIMP) is also linked with subsequent low- or high-level DNA MSI[6-8]. Methylation of the human DNA mismatch repair gene mutator L homologue 1 (hMLH1) is the principal mechanism underlying the pathogenesis of sporadic high-level MSI (MSI-H) colorectal cancer (CRC)[6] and methylation of another DNA repair gene, O-6-methylguanine DNA methyltransferase (MGMT), is linked with low-level MSI (MSI-L)[7,8]. Moreover, epigenetic inactivation of the cyclin-dependent kinase inhibitor 2A (CDKN2A/p16) by methylation has been observed in both adenomas and CRC[9,10]. These hypermethylated genes are not only probable pathogenic events in the polyp to cancer progression sequence, but are also neoplasm-specific molecular events that have the potential to be used as molecular markers for pre-malignant tumors in the colon.
In this study, we used high-sensitivity methylation-specific polymerase chain reaction (PCR) (MSP) assays for hMLH1, CDKN2A/p16, and MGMT, three genes previously shown to be aberrantly methylated in pre-malignant neoplasms in the colon[6-10]. We have applied these assays to DNA extracted from blood, normal tissue, adenomas (tubular, villous, or tubulovillous), and colon cancer of patients who underwent first time colonoscopy at the University Hospital of Ioannina, Greece. We assessed the prevalence of aberrant DNA methylation and its correlation to MSI status, as well as the temporal order and the time of their appearance during the different steps of adenoma to carcinoma progression, in an attempt to identify their possible use as molecular markers of colon carcinogenesis and furthermore their pathogenic role in the transformation of colon neoplastic adenomas to carcinoma.
The study was designed to include patients undergoing first time colonoscopy for routine clinical indications, organized in four different groups, on account of their colonoscopic and histopathological data. Patients without colonoscopic findings and histologically normal mucosa consisted group I (normal individuals - G I). Patients with tubular adenomatous polyps comprised group II (G II). Patients with polyps of increased tumorigenic potential (tubulovillous or villous adenomatous polyps) formed group III (G III), and, finally, patients with colonoscopic findings and histological evidence of CRC comprised group IV (G IV).
Seventy nine patients (44 males and 35 females, mean age: 62.5 ± 13.9 years) who underwent colonoscopy for routine clinical indications or for colon cancer screening at the Hepatogastroenterology Unit of the University Hospital of Ioannina, were included in this study, after approval from the Review Board of the University Hospital of Ioannina. Each group consisted of approximately 20 consecutive patients defined by histopathological analysis. Thus, we obtained 18 patients with tubular adenomas (10 M/8 F), 21 patients with tubulovillous or villous adenomas (12 M/9 F), and 20 colon cancer cases (11 M/9 F). Finally, 20 adenoma-free patients matched by sex and age (11 M/9 F) formed the control group.
The study was performed on freshly obtained lesions from adenomas or cancerous tissue that were resected at the time of colonoscopy. Matched normal colorectal mucosa was obtained from the resection margin that was furthest from any malignant lesion. Blood samples withdrawn at the time of colonoscopy were also included in this study. Patients with a prior history of inflammatory bowel disease, genetic CRC syndromes, or any other cancers, were excluded from our study.
DNA from the blood, fresh normal, and abnormal tissue of patients who underwent colonoscopy was extracted using the QiaAmp DNA Mini Kit (Qiagen GmbH, Hilden, Germany).
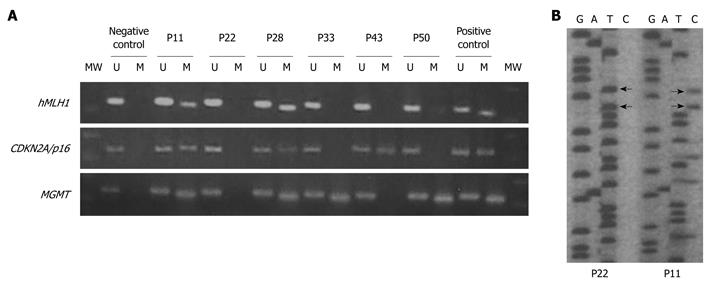
Methylation of the CpG promoter region of the three genes hMLH1, CDKN2A/p16, and MGMT was determined by MSP after bisulfite-modification of DNA samples (blood, cancerous, and non-cancerous tissue) as previously described[11,12]. These three genes have been employed in several earlier methylation studies of CIMP and CRC. In brief, the procedure was as follows: The extracted DNA underwent bisulfite modification (2 μg DNA is necessary for each experiment) using the EZ DNA methylation Gold kit (Zymo Research, Orange, CA) according to the manufacturer’s instructions. The modified DNA was used immediately for MSP or stored at -20°C for further analysis. The bisulfite-treated DNA was subject to MSP in a blinded manner using primer pairs designed to specifically amplify the methylated or unmethylated alleles for the respective genes (Table 1). Each PCR reaction mix consisted of a total volume of 50 μL containing: 3.5 mmol/L of MgCl2 (2 mmol/L for the methylated reaction); 1 × PCR Gold Buffer (Applied Biosystems, Weiterstadt, Germany); 250 μmol/L deoxynucleotide triphosphates mixture (Promega, Madison, USA); 0.1 μmol/L of forward and reverse primers (Invitrogen GmbH, Karlsruhe, Germany); 2.5 Units of AmpliTaq Gold DNA polymerase (Applied Biosystems, Weiterstadt, Germany), and the appropriate amount of bisulfite-treated DNA. The thermocycler conditions were in general as follows: 95°C for 10 min; 55 cycles of 30 s each at 95°C, specific annealing temperature for 30 s, and 1 min at 72°C; and a final extension of 10 min at 72°C. The PCR products were then subjected to horizontal gel electrophoresis on a 25 g/L agarose gel, stained with ethidium bromide, and visualized under UV transillumination. All MSP assays were repeated at least twice to validate the results. A set of known methylated and unmethylated control DNA samples was included in each round of bisulfite treatment.
| CpG status | Genes | Forward primer (5’→3’) | Reverse primer (5’→3’) | Genomic position2 | Annealing temperature (°C) | Product size (bp) |
| M | hMLH1 | ACGTAGACGTTTTATTAGGGTCGC | CCTCATCGTAACTACCCGCG | -716 to -602 | 55 | 115 |
| U | TTTTGATGTAGATGTTTTATTAGGGTTGT | ACCACCTCATCATAACTACCCACA | -721 to -598 | 55 | 124 | |
| M | MGMT1 | TTTCGACGTTCGTAGGTTTTCGC | GCACTCTTCCGAAAACGAAACG | +142 to +223 | 62 | 121 |
| U | TTTGTGTTTTGATGTTTGTAGGTTTTTGT | AACTCCACACTCTTCCAAAAACAAAACA | +137 to +230 | 62 | 133 | |
| M | CDKN2A/p16 | TTATTAGAGGGTGGGGCGGATCGC | CCACCTAAATCGACCTCCGACCG | +167 to +401 | 65 | 234 |
| U | TTATTAGAGGGTGGGGTGGATTGT | CCACCTAAATCAACCTCCAACCA | +167 to +401 | 55 | 234 |
The specificities of the MSP assays were confirmed by sequence analysis of aberrant methylation bands using the Thermo Sequenase Cy5.5 Dye Terminator Cycle Sequencing Kit (Amersham Biosciences, Piscataway, USA), in a LI-COR 4200 DNA sequencer (LI-COR Inc., USA), in parallel with the corresponding normal DNA samples, as described previously[12].
The MSI status of the 79 patients was assessed using the reference panel of five pairs of microsatellite primers: BAT25, BAT26, D2S123, D5S346, and D17S250, known as Bethesda markers[13]. PCR reactions and primer sequences have been described previously[14]. The 5’-labelled PCR products are loaded onto a 66 cm denaturing 6 mol/L urea-acrylamide gel and analyzed in a LI-COR 4200 DNA sequencer. Each PCR was run twice to ensure reproducibility of results in case the band shifts were not clearly informative in the first attempt. The MSI phenotype was identified by the presence of abnormal bands in the polyp/adenoma or carcinoma tissue DNA that were not present in blood or normal tissue DNA. MSI high (MSI-H) polyps/adenomas or carcinomas were defined as those where two of the five Bethesda markers were unstable. MSI low (MSI-L) samples were defined as a shift in only one of the five markers. Samples showing no allelic shifts were termed as MSI stable (MSS).
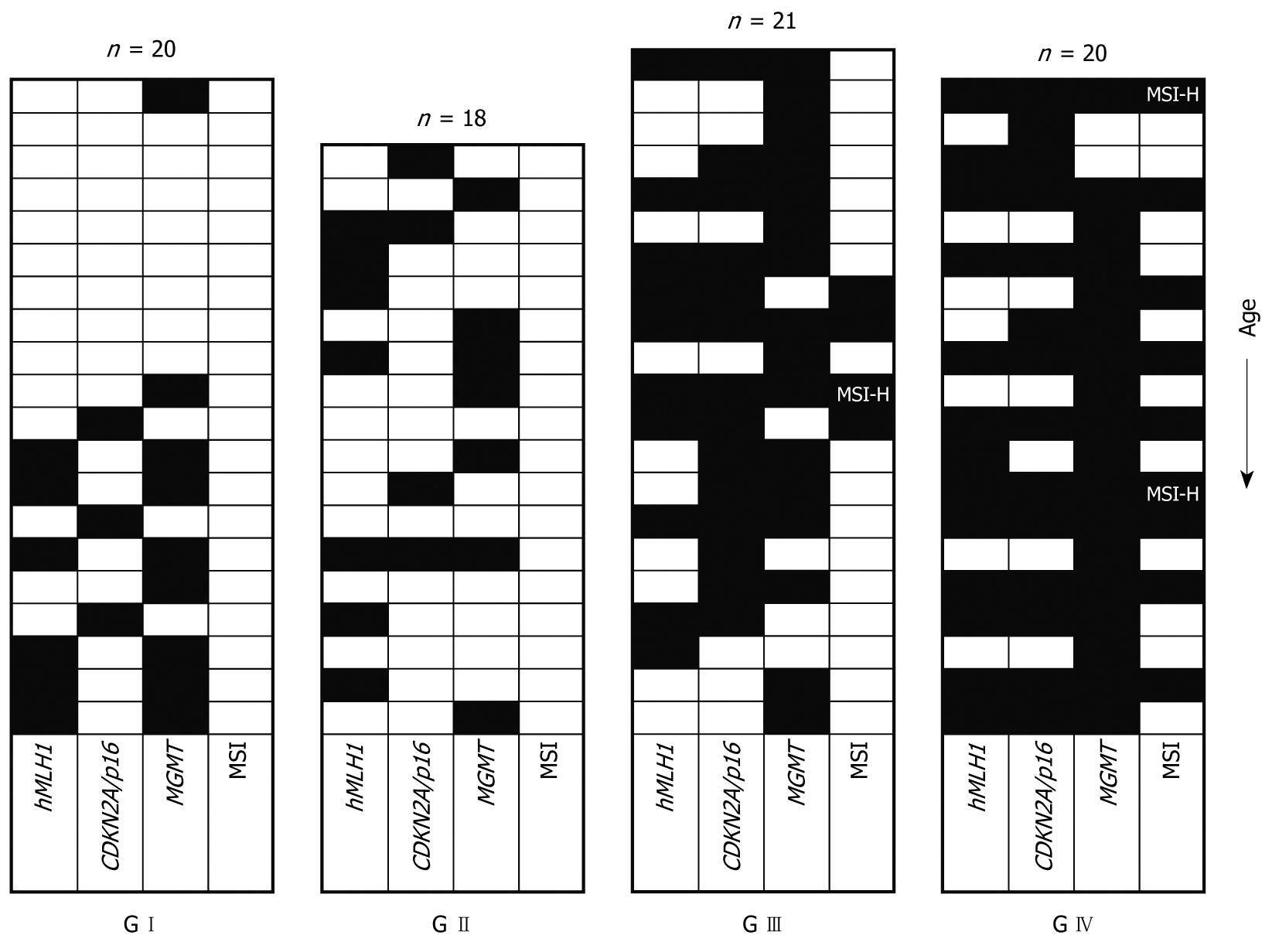
Table 2 shows the methylation patterns of the four distinct patient groups for the promoters of the genes examined. Promoter methylation was detected even in the G I group (healthy individuals). In this case, only patients aged over sixty showed methylation of some promoters (with one exception for MGMT concerning a 28-year-old woman). This indicates that methylation in normal patients is correlated to the age. By contrast,, in the patients of the G II, G III, and G IV groups, the frequency of methylation is similar in older and younger patients (> 60 years and ≤ 60 years respectively, statistical data not shown), indicating that factors other than age are probably responsible for promoter methylation in these groups (Figure 1).
| Patient group | hMLH1 (tissue) | CDKN2A/p16 (tissue) | MGMT (tissue) | MGMT (blood) | ||||
| M | U | M | U | M | U | M | U | |
| G I (n = 20) | 6/20 (30) | 14/20 (70) | 3/20 (15) | 17/20 (85) | 9/20 (46) | 11/20 (54) | 8/20 (40) | 12/20 (60) |
| G II (n = 18) | 7/18 (39) | 11/18 (61) | 4/18 (22) | 14/18 (78) | 7/18 (39) | 11/18 (61) | 1/18 (6) | 17/18 (94) |
| P = 0.563 | P = 0.581 | P = 0.666 | P = 0.0191 | |||||
| G III (n = 21) | 10/21 (48) | 11/21 (52) | 14/21 (67) | 7/21 (33) | 16/21 (76) | 5/21 (24) | 7/17 (41) | 10/17 (59) |
| P = 0.576 | P = 0.0081 | P = 0.0251 | P = 0.0191 | |||||
| G IV (n = 20) | 13/20 (65) | 7/20 (35) | 14/20 (70) | 6/20 (30) | 18/20 (90) | 2/20 (10) | 11/18 (64) | 7/18 (36) |
| P = 0.279 | P = 0.837 | P = 0.242 | P = 0.245 | |||||
Among all samples investigated, promoter methylation of hMLH1 or CDKN2A/p16 was detected only in tissue samples and not in blood (indicating that methylation of these promoters is somewhat tissue/site specific). MGMT methylation was detected in tissue as well as in blood samples. The frequency of MGMT methylation was similar in blood and tissue samples only for the G I patient group (46% in tissues vs 40% in blood, P = 0.704), as has been reported before and is also age related. However, in patient groups G II, G III, and G IV a significantly increased ratio in tissue samples vs blood samples (39% in tissues vs 6% in blood, P = 0.022; 76% vs 41%, P = 0.035 and 90% vs 64%, P = 0.043 for G II, G III and G IV, respectively) was observed. This implies that methylation of MGMT is also in part tissue/site specific. Another explanation for this could also be a low incidence of circulating cells having methylated promoters. In any case, this will require further investigation. The frequency of promoter methylation for each locus, as well as the number of methylated loci, increased from group G I to G IV (Table 2 and Figure 1), with MGMT showing a higher ratio. All of the methylated tissue samples also displayed evidence of unmethylated hMLH1, CDKN2A/p16, and MGMT, indicating that only one of the alleles is methylated or that only a part of the tissue contained cells that carried the methylated allele of the examined promoters (Figure 2).
It must be noted that hMLH1 showed similar levels of promoter methylation between groups G II and G III (39% vs 48%, P = 0.576) in contrast to CDKN2A/p16 and MGMT, which showed clearly increased levels of methylation in group G III (22% vs 67%, P = 0.008, and 39% vs 76%, P = 0.025, respectively), suggesting that hMLH1 promoter methylation is an early phenomenon in comparison to polyp formation, while methylation of CDKN2A/p16 and MGMT is correlated to the progression of polyps to more tumorigenic cases.
CDKN2A/p16 methylation was usually accompanied by methylation of another locus in tumorous and highly tumorigenic tissues (13/14 for G III and G IV), in contrast to the normal and low tumorigenic tissues (0/3 for GI and 2/4 for G II respectively). MGMT follows a rather different pattern, being in many cases the only methylated locus (5/7, 6/16, and 5/18 for G II, G III and G IV, respectively). The pattern for hMLH1 is even more complicated (Figure 1).
Of the 18 tubular adenoma patients examined 14 (77%) showed methylation in at least one of the three tested loci, while four (22%) showed no evidence of promoter methylation, and only one (6%) showed methylation in all loci (Figure 1). All 21 G III, as well as the 20 G IV patients (100%), showed promoter methylation in at least one locus. In particular, 6/21 (28%) and 11/20 (55%) for group G III and G IV, respectively, were methylated in all loci, suggesting a potentially crucial role of methylation in the progression of CRC.
According to our introductory remarks concerning the linkage between methylation status and MSI phenotype, we analysed the MSI status using the five Bethesda markers recommended by the NCI workshop[13]. Of the 79 samples examined, 66 (83%) were MSS, 10 (13%) were MSI-L and three (4%) were MSI-H (Figure 1). Only the G III and G IV patients showed MSI, which reached 14% (3/21) for MSI-L and 5% (1/21) for MSI-H for the patient group G III, whereas the corresponding percentages were 35% (7/20) and 10% (2/20) for MSI-L and MSI-H, respectively, in the G IV patient group.
Of the five Bethesda markers used for MSI analysis, BAT25 and BAT26 showed instability only in the MSI-H patients. On the other hand, the MSI-L phenotype was exclusively restricted to dinucleotide markers D2S123, D5S346, and D17S250 (3, 4 and 3 cases respectively). In the 13 cases with MSI-H or MSI-L, the following positive rates were obtained: 23% (3/13) in BAT26, 23% (3/13) in BAT25, 46% (6/13) in D2S123, 54% (7/13) in D5S346, and 46% (6/13) in D17S250, which is in accordance with previous references for cancer patients[15], as well as for patients with adenomatous polyps[16,17]. In a larger cohort of MSI-H cancer patients, Arnold et al[18] showed that the specificity of Bethesda markers was best for BAT26 and BAT25, with 99% and 95%, respectively. It is important to note that simultaneous hMLH1, CDKN2A/p16, and MGMT promoter methylation was present in 8/9 G IV patients, with the MSI phenotype as also being present in 2/4 patients of group G III. All MSI-H patients showed methylation in all loci. From the remaining three patients, the two in group G III showed methylation in two promoters (hMLH1 and CDKN2A/p16) and the one G IV patient only in the MGMT promoter. The latter case also showed instability for the D2S123 marker. In the previous work of Arnold et al[18], all Bethesda markers, except the dinucleotide repeat D2S123, had a high detection rate (up to 90%) for the combination of MSI-H cancers and hMLH1 methylation. This suggests that MSI-H cancers might originate from different pathways, e.g. one being caused by silencing the hMLH1 gene and others by as yet unrecognized mechanisms.
The transformation of normal colon epithelial cells to adenomas, and then to cancer, is believed to be an evolutionary process in which neoplastic cells acquire heritable genetic and epigenetic alterations that drive the process of carcinogenesis[19]. Gene promoter hypermethylation is increasingly recognized to play an important role in cancer development through silencing gene transcription. It is also likely that the genetic and epigenetic alterations co-operate to promote tumor formation, and that the detection of colon polyps or adenomas that present aberrant promoter methylation might identify colonic epithelium that is at significant risk of acquiring genetic alterations that will lead to colon tumor formation[20].
A number of studies have investigated the concordant methylation of multiple genes in colon adenoma[10,21,22]. Recently, aberrant promoter methylation found in different polyp forms[23,24] serves for a better understanding of aberrant CpG island methylation in the polyp/adenoma/carcinoma sequence of colorectal tumorigenesis. Thus, CIMP has been identified during several key stages in colon tumorigenesis, including aberrant crypt foci (ACF, the earliest identifiable neoplastic lesions in the colon), hyperplastic, tubular and tubulovillous/villous polyps, sporadic serrated adenomas, and tumors[3,10,22,25,26], suggesting that DNA methylation might be a pivotal event in the development of CRC. Furthermore, the aberrant methylation of MGMT and hMLH1 has been shown to silence some genes and to result in cancer promoting events, such as an MSI phenotype or k-RAS mutation[27,28].
The molecular genetics of colorectal neoplasms have been studied extensively, but few studies have addressed the clinical and pathological associations in prospectively defined patient populations. Therefore, we studied specific promoter-methylation in a cohort that underwent colonoscopy including healthy individuals, polyp-bearing patients (more or less tumorigenic), as well as CRC patients.
Although we employed the MSP assay to detect methylated hMLH1, CDKN2A/p16, and MGMT DNA in the blood samples of patients of our cohort, we were able to detect methylated DNA in blood samples only for the MGMT promoter in all patient groups, including those with CRC. We note that Grady et al[11] used MSP in a study of 20 patients, detected methylated hMLH1 DNA in the serum in 30% of patients with sporadic MSI colon cancer. This difference with our results might be attributed to the small number of MSI cancer patients of our cohort or to a different methylation site on the promoter.
We also observed increasing frequencies of promoter methylation as well as an increased number of methylated loci moving from group G I to group G IV, for the three genes examined, which is in accordance with the data of Lee et al[29]. They also observed a stepwise increase in the number and frequency of methylated genes through the stages of multistep colorectal carcinogenesis in a study including twelve loci.
Adenomas with a villous component are generally larger than tubular adenomas and have been associated with a higher risk of CRC[30-32]. A number of studies have reported higher rates of methylation in adenomas with tubulovillous or villous histology (TVAs and VAs)[10,23,24,33,34] as compared to tubular adenomas (TA). Our study also demonstrated that methylation occurs more frequently in patients with TVAs/VAs (G III) in comparison to patients with TAs for MGMT and CDKN2A/p16 (statistically significant). By contrast, for hMLH1, the two groups showed a similar ratio. An increase in MGMT methylation has been demonstrated by other studies: 38% to 61% by Petko et al[23], 30% to 65% by Kim et al[33], and 37% to 87% by Kakar et al[24]. The higher rates of MGMT methylation in our study in comparison to previous ones could be attributed in part to the use of fresh tissue samples instead of paraffin-embedded tissue samples. Our results also showed an increased frequency of CDKN2A/p16 comparable with that of Petko et al[23], who found a ratio of 10% in the methylation of CDKN2A/p16 for HPs and up to 30% for the adenomatous cases.
Furthermore, in our study, methylation, at least in one locus, reached 100% for TVAs/VAs and cancer patients, which is a higher value than that shown by Rashid et al[10].
It is likely that several different methylation pathways operate in CRC progression. Methylation of MGMT and CDKN2A/p16 loci that are found to increase from TAs to TVA/VA could be a result of the adenoma to carcinoma progression, whereas methylation of hMLH1 might be an initial step strongly associated with MSI-mediated carcinogenesis. The higher frequency of methylation in all loci for cancer patients compared to TVAs/VAs patients, as well as the increase in methylation from TAs to TVAs/VAs for MGMT and CDKN2A/p16 supports this hypothesis.
The fact that MSI is evident in TVAs/VAs and that MSI and aberrant promoter methylation are observed simultaneously, suggests that MSI and hypermethylation are dependent on each other. As it has been shown before, that among 10% to 15% of the patients with colon cancer who have MSI, approximately 70% to 80% exhibit epigenetic gene silencing of the mismatch repair gene, hMLH1[6,27,35]. Moreover, a minor fraction of MSI-L and MSS cancers also appear to be methylated at the hMLHl promoter[36]. In our study, all MSI-H patients and 9/10 MSI-L patients showed simultaneous promoter methylation of hMLH1, while the remaining one MSI-L patient showed methylation of MGMT. However, not all MSI-L patients show inactivation of MGMT by promoter methylation, as we demonstrated for two of the G III patients with the MSI-L phenotype (Figure 2).
The present study used MSP for detecting methylated alleles. MSP is a qualitative assay and does not provide quantitative information. Thus, the methylation detected by the MSP assay might not reflect gene expression, because the assay can detect only one methylated allele among 1000 unmethylated ones, and thus the vast majority of tumor cells may not harbor CpG island methylation of the given gene. The present study also shows that the methylated alleles of certain genes are present at an early stage of tumorigenesis, and that the number of genes with methylated alleles increases along the polyp/adenoma/carcinoma sequence, with MGMT being the most methylated gene.
In conclusion, our data indicates that aberrant CpG island hypermethylation occurs early and accumulates during multistep colorectal carcinogenesis, and that a temporal order exists in the methylation of tumor related genes. Furthermore, MSI is tightly connected to hMLH1 as well as to MGMT promoter methylation, indicating that inactivation and/or overloading of the mismatch repair system might have a crucial role in driving CRC progression.
Colorectal carcinogenesis is a multistep process in which the progressive accumulation of genetic and epigenetic changes leads to a malignant transformation of normal epithelial cells to adenoma and, moreover, to cancer of the colon. CpG island DNA methylation, the most extensively studied epigenetic alteration in neoplasms, is characterized by the concordant methylation of the promoter region of many tumor suppressor and DNA repair genes, such as hMLH1, CDKN2A/p16, and MGMT, although their influence on disease progression remains inconclusive.
Epigenetic changes usually begin very early in carcinogenesis, they are potentially reversible, and they can be thought of as one hit of the two-hits required for inactivation of carcinogenesis-related genes. For this reason, detection of aberrant methylation is important for early diagnosis, prognosis, and subsequent treatment of patients affected by this disease.
The frequency of promoter methylation for each promoter locus increases in the healthy tissue/adenoma/carcinoma sequence. MGMT shows the highest frequency in each group. MGMT and CDKN2A/p16 present a statistically significant increase in promoter methylation between the less and more tumorigenic form of colorectal adenomas (tubular vs tubullovillous and villous adenomas). All tubulovillous/villous adenomas bearing patients, as well as all colorectal cancer patients, showed promoter methylation in at least one of the examined loci. These findings suggest a potentially crucial role for methylation in the polyp/adenoma to cancer progression in colorectal carcinogenesis. Microsatellite instability (MSI) and methylation seem to be dependent on each other, as simultaneous hMLH1, CDKN2A/p16, and MGMT promoter methylation was present in 8/9 colorectal cancer patients showing the MSI phenotype.
The results presented here show that a series of genetic and epigenetic molecular changes drives the next step during tumorigenesis in colorectal cancers, which results in a different molecular profile for each step. This underlines the need for a detailed record of the molecular situation, in order to establish the most efficient treatment for the patient. Prospective studies supplemented by the conventional study of prognostic factors, could improve the quality and accuracy of patients’ prognosis and aid design of more efficient treatments.
Epigenetic changes: Heritable changes in gene structure that do not include the changes in DNA sequence. CpG islands: CpG rich areas located in the promoter regions of many genes. CpG island methylation: The addition of a methyl group to a cytosine residue that lies next to guanine within CpG dinucleotides. Aberrant de novo methylation of CpG islands within the promoter region might lead to silencing of gene transcription through a complex process involving chromatin condensation and histone deacetylation. MSI is a change in the length of DNA microsatellites due to the insertion or deletion of repeating units (usually 1-5 nucleotides long), caused by defects in mismatch repair genes (MLH1, MSH2, or MSH6, and others) or methylation of the MLH1 promoter.
In this study, Psofaki et al investigated promoter methylation of hMLH1, p16 and MGMT, as well as MSI, in a cohort of samples from patients underwent colonoscopy. They found accumulation of promoter methylation events during colorectal tumor progression. Promoter methylation and MSI seem to be dependent on each other. These results confirm that epigenetic inactivation is an important mechanism of tumor suppressor gene silencing. The study was well designed and the results are interesting. The manuscript is well written and includes potentially interesting findings.
Peer reviewers: Ajay Goel, PhD, Department of Internal Medicine, Division of Gastroenterology, Baylor University Medical Center and Charles A Sammons Cancer Center, 3500 Gaston Avenue, Suite H-250, Dallas, TX 75246, United States; Lin Zhang, PhD, Associate Professor, Department of Pharmacology and Chemical Biology, University of Pittsburgh Cancer Institute, University of Pittsburgh School of Medicine, UPCI Research Pavilion, Room 2.42d, Hillman Cancer Center, 5117 Centre Ave., Pittsburgh, PA 15213-1863, United States; Sung-Gil Chi, Professor, School of Life Sciences and Biotechnology, Korea University, #301, Nok-Ji Building, Seoul 136-701, South Korea
S- Editor Wang JL L- Editor Stewart GJ E- Editor Lin YP
| 1. | Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558-561. [Cited in This Article: ] |
| 2. | Perucho M. Tumors with microsatellite instability: many mutations, targets and paradoxes. Oncogene. 2003;22:2223-2225. [Cited in This Article: ] |
| 3. | Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681-8686. [Cited in This Article: ] |
| 4. | Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141-196. [Cited in This Article: ] |
| 5. | Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988-993. [Cited in This Article: ] |
| 6. | Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870-6875. [Cited in This Article: ] |
| 7. | Jass JR, Biden KG, Cummings MC, Simms LA, Walsh M, Schoch E, Meltzer SJ, Wright C, Searle J, Young J. Characterisation of a subtype of colorectal cancer combining features of the suppressor and mild mutator pathways. J Clin Pathol. 1999;52:455-460. [Cited in This Article: ] |
| 8. | Whitehall VL, Walsh MD, Young J, Leggett BA, Jass JR. Methylation of O-6-methylguanine DNA methyltransferase characterizes a subset of colorectal cancer with low-level DNA microsatellite instability. Cancer Res. 2001;61:827-830. [Cited in This Article: ] |
| 9. | Guan RJ, Fu Y, Holt PR, Pardee AB. Association of K-ras mutations with p16 methylation in human colon cancer. Gastroenterology. 1999;116:1063-1071. [Cited in This Article: ] |
| 10. | Rashid A, Shen L, Morris JS, Issa JP, Hamilton SR. CpG island methylation in colorectal adenomas. Am J Pathol. 2001;159:1129-1135. [Cited in This Article: ] |
| 11. | Grady WM, Rajput A, Lutterbaugh JD, Markowitz SD. Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer Res. 2001;61:900-902. [Cited in This Article: ] |
| 12. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [Cited in This Article: ] |
| 13. | Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248-5257. [Cited in This Article: ] |
| 14. | Loukola A, Eklin K, Laiho P, Salovaara R, Kristo P, Järvinen H, Mecklin JP, Launonen V, Aaltonen LA. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC). Cancer Res. 2001;61:4545-4549. [Cited in This Article: ] |
| 15. | Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Rüschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749-4756. [Cited in This Article: ] |
| 16. | Iino H, Jass JR, Simms LA, Young J, Leggett B, Ajioka Y, Watanabe H. DNA microsatellite instability in hyperplastic polyps, serrated adenomas, and mixed polyps: a mild mutator pathway for colorectal cancer? J Clin Pathol. 1999;52:5-9. [Cited in This Article: ] |
| 17. | Ricciardiello L, Goel A, Mantovani V, Fiorini T, Fossi S, Chang DK, Lunedei V, Pozzato P, Zagari RM, De Luca L. Frequent loss of hMLH1 by promoter hypermethylation leads to microsatellite instability in adenomatous polyps of patients with a single first-degree member affected by colon cancer. Cancer Res. 2003;63:787-792. [Cited in This Article: ] |
| 18. | Arnold CN, Goel A, Compton C, Marcus V, Niedzwiecki D, Dowell JM, Wasserman L, Inoue T, Mayer RJ, Bertagnolli MM. Evaluation of microsatellite instability, hMLH1 expression and hMLH1 promoter hypermethylation in defining the MSI phenotype of colorectal cancer. Cancer Biol Ther. 2004;3:73-78. [Cited in This Article: ] |
| 19. | Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet. 2002;3:101-128. [Cited in This Article: ] |
| 20. | Esteller M. Epigenetic lesions causing genetic lesions in human cancer: promoter hypermethylation of DNA repair genes. Eur J Cancer. 2000;36:2294-2300. [Cited in This Article: ] |
| 21. | Bariol C, Suter C, Cheong K, Ku SL, Meagher A, Hawkins N, Ward R. The relationship between hypomethylation and CpG island methylation in colorectal neoplasia. Am J Pathol. 2003;162:1361-1371. [Cited in This Article: ] |
| 22. | Park SJ, Rashid A, Lee JH, Kim SG, Hamilton SR, Wu TT. Frequent CpG island methylation in serrated adenomas of the colorectum. Am J Pathol. 2003;162:815-822. [Cited in This Article: ] |
| 23. | Petko Z, Ghiassi M, Shuber A, Gorham J, Smalley W, Washington MK, Schultenover S, Gautam S, Markowitz SD, Grady WM. Aberrantly methylated CDKN2A, MGMT, and MLH1 in colon polyps and in fecal DNA from patients with colorectal polyps. Clin Cancer Res. 2005;11:1203-1209. [Cited in This Article: ] |
| 24. | Kakar S, Deng G, Cun L, Sahai V, Kim YS. CpG island methylation is frequently present in tubulovillous and villous adenomas and correlates with size, site, and villous component. Hum Pathol. 2008;39:30-36. [Cited in This Article: ] |
| 25. | Chan AO, Broaddus RR, Houlihan PS, Issa JP, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol. 2002;160:1823-1830. [Cited in This Article: ] |
| 26. | Wynter CV, Walsh MD, Higuchi T, Leggett BA, Young J, Jass JR. Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut. 2004;53:573-580. [Cited in This Article: ] |
| 27. | Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL, Sedwick WD. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci USA. 1998;95:8698-8702. [Cited in This Article: ] |
| 28. | Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427-5440. [Cited in This Article: ] |
| 29. | Lee S, Hwang KS, Lee HJ, Kim JS, Kang GH. Aberrant CpG island hypermethylation of multiple genes in colorectal neoplasia. Lab Invest. 2004;84:884-893. [Cited in This Article: ] |
| 30. | Atkin WS, Morson BC, Cuzick J. Long-term risk of colorectal cancer after excision of rectosigmoid adenomas. N Engl J Med. 1992;326:658-662. [Cited in This Article: ] |
| 31. | Loeve F, van Ballegooijen M, Snel P, Habbema JD. Colorectal cancer risk after colonoscopic polypectomy: a population-based study and literature search. Eur J Cancer. 2005;41:416-422. [Cited in This Article: ] |
| 32. | Martínez ME, Sampliner R, Marshall JR, Bhattacharyya AK, Reid ME, Alberts DS. Adenoma characteristics as risk factors for recurrence of advanced adenomas. Gastroenterology. 2001;120:1077-1083. [Cited in This Article: ] |
| 33. | Kim HC, Roh SA, Ga IH, Kim JS, Yu CS, Kim JC. CpG island methylation as an early event during adenoma progression in carcinogenesis of sporadic colorectal cancer. J Gastroenterol Hepatol. 2005;20:1920-1926. [Cited in This Article: ] |
| 34. | Kim YH, Petko Z, Dzieciatkowski S, Lin L, Ghiassi M, Stain S, Chapman WC, Washington MK, Willis J, Markowitz SD. CpG island methylation of genes accumulates during the adenoma progression step of the multistep pathogenesis of colorectal cancer. Genes Chromosomes Cancer. 2006;45:781-789. [Cited in This Article: ] |
| 35. | Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808-811. [Cited in This Article: ] |
| 36. | Laiho P, Launonen V, Lahermo P, Esteller M, Guo M, Herman JG, Mecklin JP, Järvinen H, Sistonen P, Kim KM. Low-level microsatellite instability in most colorectal carcinomas. Cancer Res. 2002;62:1166-1170. [Cited in This Article: ] |