Published online Aug 28, 2008. doi: 10.3748/wjg.14.5008
Revised: July 26, 2008
Accepted: August 2, 2008
Published online: August 28, 2008
AIM: To explore the existence and distribution of prohibitin (PHB) in nuclear matrix and its co-localization with products of some related genes during the differentiation of human hepatocarcinoma SMMC-7721 cells.
METHODS: The nuclear matrix of the SMMC-7721 cells cultured with or without 5 × 10-3 mmol/L hexamethylene bisacetamide (HMBA) was selectively extracted. Western blot was used to analyze the expression of PHB in nuclear matrix; immunofluorescence microscope observation was used to analyze the distribution of PHB in cell. LCSM was used to observe the co-localization of PHB with products of oncogenes and tumor suppressor genes.
RESULTS: Western blot analysis showed that PHB existed in the composition of nuclear matrix proteins and was down-regulated by HMBA treatment. Immunofluorescence observation revealed that PHB existed in the nuclear matrix, and its distribution regions and expression levels were altered after HMBA treatment. Laser scanning confocal microscopy revealed the co-localization between PHB and the products of oncogenes or tumor repression genes including c-fos, c-myc, p53 and Rb and its alteration of distributive area in the cells treated by HMBA.
CONCLUSION: These data confirm that PHB is a nuclear matrix protein, which is located in the nuclear matrix, and the distribution and expression of PHB and its relation with associated genes may play significant roles during the differentiation of SMMC-7721 cells.
- Citation: Xu DH, Tang J, Li QF, Shi SL, Chen XF, Liang Y. Positional and expressive alteration of prohibitin during the induced differentiation of human hepatocarcinoma SMMC-7721 cells. World J Gastroenterol 2008; 14(32): 5008-5014
- URL: https://www.wjgnet.com/1007-9327/full/v14/i32/5008.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.5008
| Antibodies | Source | Antibodies | Source |
| Mouse anti-human PHB | NeoMarkers Co. | Goat anti-mouse IgG/HRP | Boster Co. |
| Goat anti-rabbit IgG-FITC | KPL Co. | Goat anti-rabbit IgG/HRP | Boster Co. |
| Goat anti-mouse IgG-TRITC | KPL Co. | Rabbit anti-p53 | Boster Co. |
| Rabbit anti-human β-actin | Boster Co. | Rabbit anti-pRb | Boster Co. |
| Rabbit anti-c-Fos | Boster Co. | Rabbit anti-c-Myc | Boster Co. |
Prohibitin (PHB) is a tumor suppressor protein that is expressed in a variety of cell lines. It is not only localized to the inner membrane of mitochondria, where it acts as a chaperone protein, but is also present in the nucleus where it negatively regulates transcription. PHB plays important roles in the regulation of cell growth, proliferation, differentiation, aging and apoptosis. It is also involved in the genesis of tumor and some degenerative diseases[1-3]. In cell differentiation, some studies have found that the expression level of PHB was very low in rapidly proliferating cells, whereas it was much higher in cells undergoing differentiation. This indicates that PHB may promote cell differentiation and suppress cell proliferation[4]. Heretofore, over-expression of PHB has been found in various tumor cells. This seems to be conflictive with its tumor suppressive function. So far, the mechanism of its subcellular localization, nuclear transportation and regulation of cell proliferation and differentiation are not well understood. Our previous studies revealed that PHB existed in nuclear matrix extractions of human adenocarcinoma MGc80-3 cells, and its expression level was altered during the differentiation induced by hexamethylene bisacetamide (HMBA)[5]. Furthermore, in the differentiation of human osteosarcoma MG-63 cells, we observed similar results[6]. This implies that PHB might be a common differentially expressed nuclear matrix protein in some tumor cells. In this study, we further studied the existence, localization, expression alteration of PHB in the nuclear matrix and the relationship between PHB and related products of oncogenes during the differentiation of human hepatocarcinoma SMMC-7721 cells induced by HMBA. This study provides scientific evidence for the function of PHB during cell differentiation and understanding of the mechanism of development of cancer and its reversion.
Human hepatocarcinoma SMMC-7721 cells were obtained from China Center for Type Culture Collection (Wuhan University). RPMI-1640 was from Gibco Co., newborn calf serum was from Hangzhou Sijiqing Biological Engineering Materials Co., Ltd. HMBA was from Sigma Co. The antibodies used are listed in Table 1.
Human hepatocarcinoma SMMC-7721 cells were cultured at 37°C in RPMI-1640 medium (pH 7.2) supplemented with 15% newborn calf serum, 100 U/mL penicillin, 100 U/mL streptomycin and 50 μg/mL kanamycin. Cells were passaged in log phase. Twenty-four hours after passaging the cells the experimental group was treated with culture media containing 5 mmol/L HMBA. Fresh culture media was added to the cells every 48-72 h. Cells were harvested at subconfluency.
The cells were selectively extracted as described in our previous article[7]. The nuclear matrix-intermediate filament (NM-IF) samples on the cover slip strips after the selective extraction were prefixed in 2% glutaraldehyde at 4°C for 30 min, and rinsed in phosphate-buffered saline (PBS), pH 7.4. The samples were then stained with 0.2% Coomassie brilliant blue for 20 min, washed in distilled water, air dried, clarified by xylene, enveloped in the resin, and observed using an Olympus BH-2 microscope.
Nuclear matrix proteins of SMMC-7721 cells were purified by using a routine method with a few improvements[8]. The SMMC-7721 cells were washed with PBS and extracted with cytoskeleton buffer (CSK100) (10 mmol/L PIPES pH 6.8, 300 mmol/L sucrose, 100 mmol/L NaCl, 4 mmol/L CaCl2, 1.0 mmol/L PMSF, 0.5% Triton X-100) at 0°C for 10 min, and subjected to centrifugation for 5 min at 400 g. The pellet was washed twice with CSK50 (10 mmol/L PIPES pH 6.8, 300 mmol/L sucrose, 50 mmol/L NaCl, 4 mmol/L CaCl2, 1.0 mmol/L PMSF, 0.5% Triton X-100) and digested for 30 min at 25°C in the same buffer containing 300 U/mL DNase I. One mole per liter ammonium sulfate was added dropwise to a final concentration of 0.25 mmol/L. After incubation for 15 min, the nuclear matrix proteins were pelleted by centrifugation at 1000 r/min for 5 min, and washed once with the CSK50 buffer, then stored at -80°C.
The nuclear matrix proteins were separated by SDS-PAGE and then transferred onto PVDF membranes. Nonspecific reactivity was blocked by incubation at room temperature for 1.5 h in 5% BSA buffer. The membrane was then incubated with PHB (1:2000) primary antibody at room temperature for 2 h. After washing, a horseradish peroxidase tagged secondary antibody was used to detect the primary antibody. Reactive protein was detected by the enhanced chemiluminescence (ECL) detection system (Pierce). As a negative control, the primary antibody was replaced by 5% BSA buffer. β-actin was also detected as an internal control.
The NM-IF samples on the cover slip were prefixed in 4% paraformaldehyde at 4°C for 10 min, rinsed in the TPBS (contain 0.5% Triton X-100) twice, 5 min each, blocked by 5% BSA at room temperature for 1 h, incubated with mouse anti-PHB monoclonal antibody (1:300) at room temperature for 30 min, and 4°C overnight, then washed 3 times with TPBS. Then, the cells were incubated with goat anti-mouse secondary antibody labeled with fluorescence dye TRITC, washed with water and dried by airing. Afterwards, 90% glycerol/PBS was applied and the cells observed using fluorescence microscopy. The entire process after incubation with secondary antibody was performed in the dark. The primary antibody was replaced by 5% BSA buffer as a negative control.
Cells on the cover slips were rinsed in PBS, submerged in TBS (including 0.5% Triton X-100) for 30 min. After being washed with PBS, the cells were fixed in 4% paraformaldehyde for 10 min, blocked with 5% BSA at room temperature for 1 h, and then incubated with dual primary antibodies at room temperature for 30 min and then 4°C overnight. The dual primary antibody sets comprised of PHB (1:1000)/c-Fos (1:200), PHB (1:1000)/c-Myc (1:200), PHB (1:1000)/p53 (1:200), PHB (1:1000)/pRb (1:200). After being washed with TTBS, the cells were incubated with different secondary antibody sets (goat anti rabbit or goat anti mouse) which were labeled with FITC and TRITC, respectively, at room temperature for 30 min and then 4°C overnight, washed with TPBS, enveloped with 90% glycerol/PBS after drying and then observed under LSCM (TCS- SP2 MP).
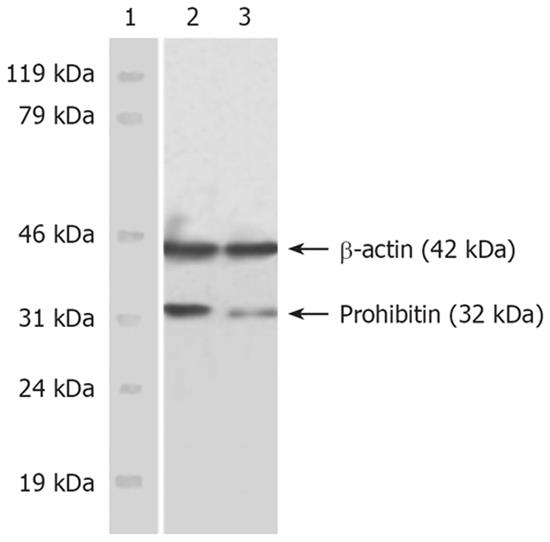
Western blot results showed the dominant PHB band located at 32 kDa (PHB) and β-actin positioned at 42 kDa. The immunobead of PHB in the nuclear matrix of SMMC-7721 cells was obviously fuscous, while it was light and thin in HMBA-treated cells. The expression level of hnRNP A2/B1 was significantly decreased. The expression level of β-actin, which was referred to as endogenous control, had no obvious changes (Figure 1).
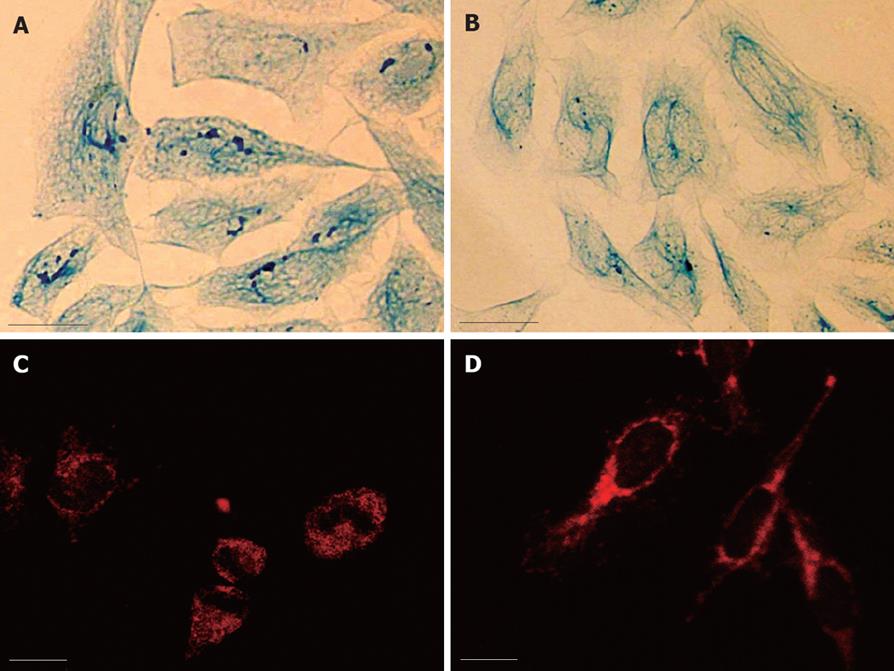
Light microscopy observation revealed that the intermediate filaments in SMMC-7721 cells were sparse and arranged irregularly. In HMBA-treated MG-63 cells, the whole framework became more outspread, and the NM-IF system showed characteristics of uniform distribution. The intermediate filaments, which were uniformly stained, spread from the region around the nucleus to the cellular edge and formed a well-distributed and regular network throughout the cytoplasmic region. The nuclear matrix filaments were abundant and evenly distributed (Figure 2A and B).
The observation of immunofluorescence revealed the localization and expression of PHB. PHB was labeled with TRITC (red). The results showed PHB existed in the whole cell, but was weak in nuclei where it was scattered as little particles. It was relatively strong in regions near the nuclear membrane. The distribution of PHB was uniform in the cytoplasmic region (Figure 2C). After treatment with HMBA, the distribution and expression of PHB in the NM-IF system was significantly altered. The holistic intensity of the fluorescence in nuclear matrix and nuclear lamina region dropped dramatically, while the fluorescence in the cytoplasmic region of HMBA-treated cells strengthened. It displayed a tendency of transferring from NM to lamina and cytoplasm of PHB (Figure 2D).
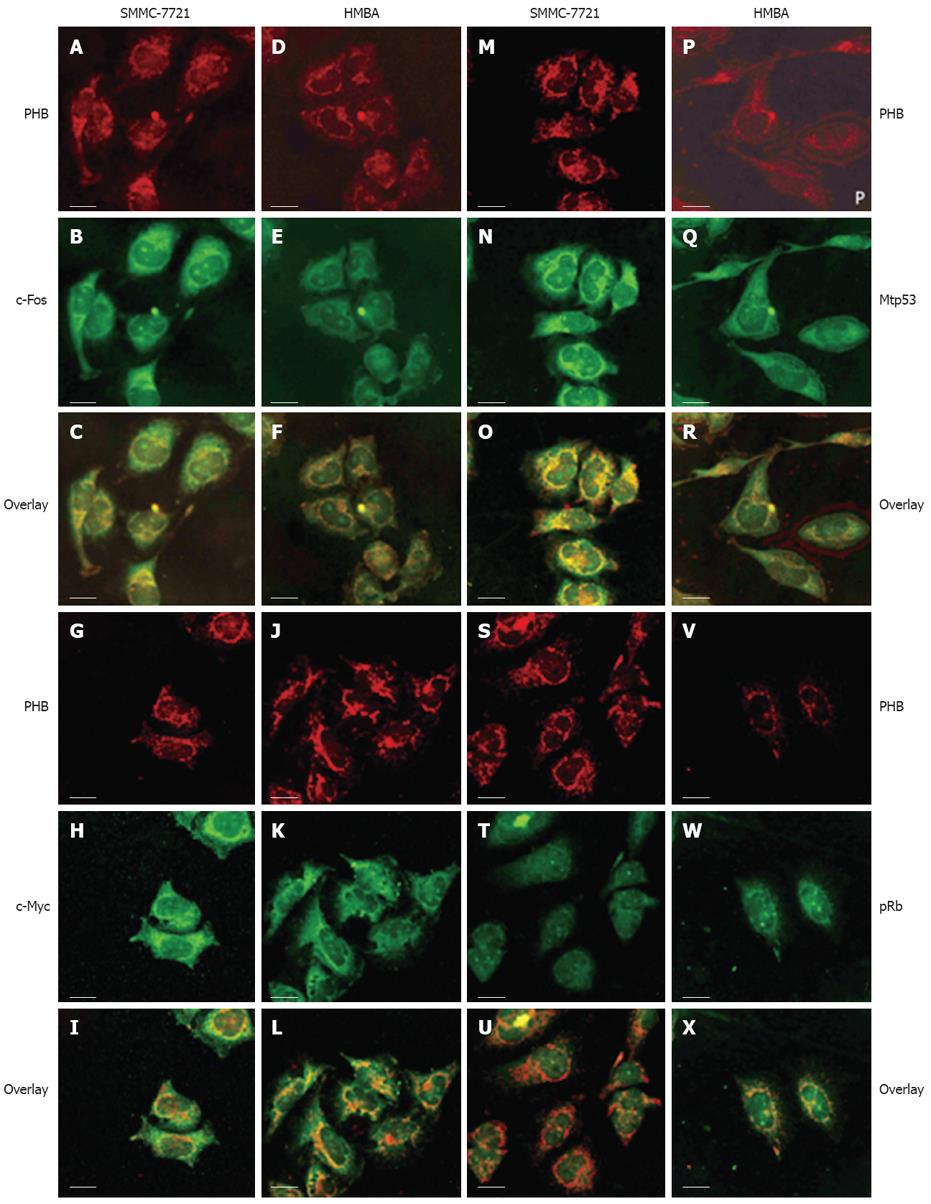
The localization of PHB and the opposite proteins including c-Fos, c-Myc, p53 and pRb were observed by LSCM. PHB was labeled with TRITC (red), other proteins were labeled with FITC (green). The co-localization fluorescence was yellow or orange when two different colors of fluorescence overlapped (Figure 3).
In SMMC-7721 cells, PHB mainly distributed in the nucleus, nuclear membrane, cytoplasm and the edge region of cells. In karyoplasm, the fluorescence was relatively weak and scattered unevenly. In SMMC-7721 cells treated with HMBA, PHB expression became decreased or even disappeared in the nuclear regions and was enhanced and scattered broadly in the cytoplasm.
In SMMC-7721 cells, c-Fos was distributed mainly in the nucleolus region. The yellow overlapped fluorescence indicated co-localization between PHB and c-Fos existed mainly in the nucleolus region. In SMMC-7721 cells treated with HMBA, the holistic intensity of PHB and c-Fos fluorescence all weakened. The overlapped fluorescence indicated that the co-localization between PHB and c-Fos in the nucleolus weakened, but the co-localized fluorescence in karyotheca and cytoplasm was enhanced. It seemed that the co-localized region of both proteins transferred from the nucleolus region to karyotheca and cytoplasm (Figure 3A-F).
In SMMC-7721 cells, the highly-expressed c-Myc distributed mainly in the nucleolus and its peripheral regions. The overlapped fluorescence indicated they were co-localized in karyoplasms and karyotheca. In SMMC-7721 cells induced by HMBA, c-Myc dispersed in the whole cell, and its expression decreased. The overlapped fluorescence of PHB and c-Myc showed that the co-localization of two proteins strengthened in the karyotheca region, and clustered co-localization fluorescence could be seen in some special places. It indicated the tendency of transportation from karyoplasm to the nearby nuclear membrane (Figure 3G-L).
Due to the short half-life of wild-p53 in tumor cells (6 min), the p53 detected in this article was mostly its mutant counterpart namely mtp53. In SMMC-7721 cells, the highly-expressed mtp53 concentrated mainly in the nucleolus and evenly distributed in other regions of the nucleus. The overlapped fluorescence indicated the co-localization distributed mainly in the region of karyoplasm and cytoplasm. In SMMC-7721 cells induced by HMBA, mtP53 decreased and its distribution was much more dispersive than that of untreated cells. The overlapped fluorescence showed the co-localization region of two proteins existed in the cytoplasm and lamina regions, and the co-localization fluorescence weakened remarkably after treatment (Figure 3M-R).
In SMMC-7721 cells, Rb expression in the nucleolus was stronger than in other regions where it was relatively weak and distributed evenly. The overlapped fluorescence indicated they were co-localized in most regions of the nuclear membrane and cytoplasm. After treatment with HMBA, Rb increased remarkably and localized mainly at the nuclear membrane and nucleus regions after HMBA treatment. The overlapped fluorescence indicated the co-localization region of two proteins existed mainly in the nuclear membrane. It also showed their co-localization had a tendency of transferring from the cytoplasm to the nuclear membrane during the differentiation of SMMC-7721 cells (Figure 3S-X).
PHB is an important tumor suppressor protein. It not only mediates signaling of cell proliferation by acting as a membrane receptor, but is also involved in the maintenance of stability and function of mitochondria. Previous studies have shown that PHB was mainly present in the mitochondria, and could be found in other subcellular organelles rarely[9,10]. So far, the localization of PHB in the nuclear matrix has not been reported. In this study, Western blot analysis confirmed that PHB was one of the components of the nuclear matrix and its expression in the nuclear matrix was down-regulated remarkably after treatment with HMBA. Immunofluorescence microscopy revealed that PHB was distributed mainly in the regions of the nuclear membrane and cytoplasm of SMMC-7721 cells. These results indicate that PHB exists not only in the cytoplasm, but also in the nuclear matrix of SMMC-7721 cells. Therefore, our laboratory, for the first time, has discovered the subcellular location of PHB in the nuclear matrix and showed it was a nuclear matrix protein.
PHB has great relevance to the development of cancer. Recent studies have shown that expression of PHB was abnormal in a variety of tumor cell lines[11-14]. In this study, using Western blot analysis, we found PHB is expressed highly in the nuclear matrix of SMMC-7721 cells, but in cells treated with HMBA, its expression was down-regulated. Immunofluorescent microscopy showed that the distribution of PHB in the nuclear matrix of SMMC-7721 cells was altered after HMBA treatment. PHB was mainly distributed in the karyoplasms and cytoplasm region, while in the HMBA-treated cells, PHB localized to the nuclear membrane and cytoplasm. PHB might have undergone translocation from nucleus to cytoplasm. Over expression of PHB has been reported in many kinds of tumor cells[15,16]. Our previous studies on the nuclear matrix proteins of human adenocarcinoma MGc80-3 and human osteosarcoma MG-63 cells showed that PHB existed as a component of the nuclear matrix, and expression of PHB was down-regulated during differentiation. Therefore, our results further affirmed the alterations of PHB expression, and confirmed the involvement of PHB in the regulation of tumor cell proliferation and differentiation.
The regulation of PHB in cell proliferation, senescence, apoptosis and differentiation involves complicated molecular mechanisms, including interaction between PHB and associated regulators in signaling and cell cycle, the subcellular localization of PHB in the cell, the phosphorylation level of PHB, and the connection between PHB and related oncogenes[17-19]. In this study, the results of laser scanning confocal microscopy revealed that there was a co-localizational relationship between PHB and the products of c-fos, c-myc, p53 and Rb genes in SMMC-7721 cells, and the co-localization areas changed after HMBA treatment.
c-fos and c-myc are oncogenes whose expressions are up-regulated frequently in hepatocellular carcinomas (HCC). Our research indicated PHB colocalized with c-Fos, c-Myc in the nuclear region of SMMC-7721 cells. When SMMC-7721 cells were induced into differentiation, the co-localization relationship of PHB with c-Fos and c-Myc strengthened in the regions of lamina and karyotheca, but weakened in the karyoplasms. Other studies showed that PHB was a downstream target gene of c-myc and c-myc could bind to the promoter region of the PHB gene[20]. This indicated the close relation between PHB and c-Myc. The direct interaction of PHB and c-Fos has not yet been reported. Our research showed the co-localization between PHB and c-Myc, c-Fos in the region of the nucleus, and its alteration in SMMC-7721 cells after treatment with HMBA. It suggested that PHB might interact directly with the products of c-myc and c-fos genes. Our study discovered that PHB and p53 were co-localized at the nuclear membrane and karyoplasms region. The co-localization fluorescence transferred from nucleus to cytoplasm after HMBA treatment. This suggested that PHB might have direct connection with p53. Recently, some studies showed that PHB and p53 could cooperate with each other and participated in regulating downstream gene expression. Some studies found co-localization between PHB, p53 and E2F1 in the nucleus of breast cancer cells, and demonstrated PHB could activate transcription mediated by p53 and enhance binding of p53 with the promoter. It is also reported that p53-PHB complex translocated from nucleus to cytoplasm after stimulation by the signal of apoptosis[21-23]. Our results were consistent with former studies. It implies that PHB can directly interact with p53 and participate in the regulation of transcription mediated by p53. The co-transferring of p53 and PHB may be one of the mechanisms that mediate regulation of cell proliferation and differentiation. The studies about Rb and PHB showed that PHB could cooperate with products of Rb, a dominant tumor suppressor gene which regulates transcriptional activity of E2F. PHB could also form a triad with Rb and E2F in the nucleus, suppress activity of E2F, and thereby inhibit cell proliferation[24,25]. The results of this study revealed that the co-localizational fluorescence in the region of the nuclear lamina was enhanced after HMBA treatment. It is obvious that the alteration is correlative with the state of proliferation and differentiation of SMMC-7721 cells. Nuclear lamina is the active site of DNA replication and transcription. Because of these, the alteration of PHB transferring and its enhanced combination with Rb in the lamina might repress the activity of E2F which promotes transcription of its downstream genes.
Taken together, these findings suggest that the subcellular localization and altered expression of PHB may have a potential role in the differentiation and phenotype reversion of SMMC-7721 cells by cooperating with products of oncogenes or tumor suppressor genes. Further investigation of the function of PHB in the nuclear matrix will help to clarify the mechanism of cell differentiation, cell cancer development and its reversion.
Previous studies showed that prohibitin (PHB) played important roles in the regulation of cell growth, proliferation, differentiation and tumorigenesis. However, the existence and expression of PHB in the nuclear matrix of tumor cells have not been well illustrated, and the functions of PHB in the induced differentiation of human hepatocarcinoma SMMC-7721 cells have also not been investigated in detail.
To identify differentially expressed nuclear matrix proteins and analyze their function in cancer cell differentiation is one of the most interesting hotspots in current studies of nuclear matrix.
The authors revealed for the first time that PHB was a nuclear matrix protein in SMMC-7721 cells, and the distribution/expression of PHB and its relation with associated genes played a significant roles during the differentiation of SMMC-7721 cells.
PHB can be used as a potential target in both diagnosis and treatment of tumors.
Nuclear matrix is the residual protein structure that remains after the nuclei are depleted of the nuclear membranes, histones, soluble nuclear proteins and nucleic acids. The structures that remain in matrix preparations are the nuclear lamina, the residual nucleolus and the fibrillogranular network.
The study is about the expression and distribution of proteins. Scientific content is interesting as several protein complexes are involved in cellular processes and identification of a role for PHB in carcinogenesis by association with other oncogenes is intriguing.
Peer reviewer: Devanshi Seth, PhD, Drug Health Services & Centenary Institute, Royal Prince Alfred Hospital, Missenden Road, Camperdown NSW 2050, Australia
S- Editor Li DL L- Editor Rippe RA E- Editor Ma WH
| 1. | Mishra S, Murphy LC, Nyomba BL, Murphy LJ. Prohibitin: a potential target for new therapeutics. Trends Mol Med. 2005;11:192-197. [Cited in This Article: ] |
| 2. | Dell’Orco RT, McClung JK, Jupe ER, Liu XT. Prohibitin and the senescent phenotype. Exp Gerontol. 1996;31:245-252. [Cited in This Article: ] |
| 3. | McClung JK, Jupe ER, Liu XT, Dell’Orco RT. Prohibitin: potential role in senescence, development, and tumor suppression. Exp Gerontol. 1995;30:99-124. [Cited in This Article: ] |
| 4. | Dixit VD, Sridaran R, Edmonsond MA, Taub D, Thompson WE. Gonadotropin-releasing hormone attenuates pregnancy-associated thymic involution and modulates the expression of antiproliferative gene product prohibitin. Endocrinology. 2003;144:1496-1505. [Cited in This Article: ] |
| 5. | Zhao CH, Li QF. Altered profiles of nuclear matrix proteins during the differentiation of human gastric mucous adenocarcinoma MGc80-3 cells. World J Gastroenterol. 2005;11:4628-4633. [Cited in This Article: ] |
| 6. | Li QF, Zhao CH, Tang J, Liu QR, Shi SL. [Localization of prohibitin in nuclear matrix and its expressive alteration during the differentiation of human osteosarcoma MG-63 cells induced by HMBA]. Fen Zi Xi Bao Sheng Wu Xue Bao. 2008;41:1-10. [Cited in This Article: ] |
| 7. | Li QF. Effect of retinoic acid on the changes of nuclear matrix in termediate filament system in gastric carcinoma cells. World J Gastroenterol. 1999;5:417-420. [Cited in This Article: ] |
| 8. | Michishita E, Kurahashi T, Suzuki T, Fukuda M, Fujii M, Hirano H, Ayusawa D. Changes in nuclear matrix proteins during the senescence-like phenomenon induced by 5-chlorodeoxyuridine in HeLa cells. Exp Gerontol. 2002;37:885-890. [Cited in This Article: ] |
| 9. | Artal-Sanz M, Tsang WY, Willems EM, Grivell LA, Lemire BD, van der Spek H, Nijtmans LG. The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J Biol Chem. 2003;278:32091-32099. [Cited in This Article: ] |
| 10. | Nijtmans LG, Artal SM, Grivell LA, Coates PJ. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell Mol Life Sci. 2002;59:143-155. [Cited in This Article: ] |
| 11. | Jang JS, Cho HY, Lee YJ, Ha WS, Kim HW. The differential proteome profile of stomach cancer: identification of the biomarker candidates. Oncol Res. 2004;14:491-499. [Cited in This Article: ] |
| 12. | Hussain-Hakimjee EA, Peng X, Mehta RR, Mehta RG. Growth inhibition of carcinogen-transformed MCF-12F breast epithelial cells and hormone-sensitive BT-474 breast cancer cells by 1alpha-hydroxyvitamin D5. Carcinogenesis. 2006;27:551-559. [Cited in This Article: ] |
| 13. | Tsai HW, Chow NH, Lin CP, Chan SH, Chou CY, Ho CL. The significance of prohibitin and c-Met/hepatocyte growth factor receptor in the progression of cervical adenocarcinoma. Hum Pathol. 2006;37:198-204. [Cited in This Article: ] |
| 14. | Qi Y, Chiu JF, Wang L, Kwong DL, He QY. Comparative proteomic analysis of esophageal squamous cell carcinoma. Proteomics. 2005;5:2960-2971. [Cited in This Article: ] |
| 15. | Campbell IG, Allen J, Eccles DM. Prohibitin 3‘ untranslated region polymorphism and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:1273-1274. [Cited in This Article: ] |
| 16. | Manjeshwar S, Branam DE, Lerner MR, Brackett DJ, Jupe ER. Tumor suppression by the prohibitin gene 3’untranslated region RNA in human breast cancer. Cancer Res. 2003;63:5251-5256. [Cited in This Article: ] |
| 17. | Tatsuta T, Model K, Langer T. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol Biol Cell. 2005;16:248-259. [Cited in This Article: ] |
| 18. | Wang S, Nath N, Adlam M, Chellappan S. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene. 1999;18:3501-3510. [Cited in This Article: ] |
| 19. | Coates PJ, Nenutil R, McGregor A, Picksley SM, Crouch DH, Hall PA, Wright EG. Mammalian prohibitin proteins respond to mitochondrial stress and decrease during cellular senescence. Exp Cell Res. 2001;265:262-273. [Cited in This Article: ] |
| 20. | Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA. 2002;99:6274-6279. [Cited in This Article: ] |
| 21. | Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem. 2003;278:47853-47861. [Cited in This Article: ] |
| 22. | Rastogi S, Joshi B, Fusaro G, Chellappan S. Camptothecin induces nuclear export of prohibitin preferentially in transformed cells through a CRM-1-dependent mechanism. J Biol Chem. 2006;281:2951-2959. [Cited in This Article: ] |
| 23. | Joshi B, Rastogi S, Morris M, Carastro LM, DeCook C, Seto E, Chellappan SP. Differential regulation of human YY1 and caspase 7 promoters by prohibitin through E2F1 and p53 binding sites. Biochem J. 2007;401:155-166. [Cited in This Article: ] |
| 24. | Wang S, Fusaro G, Padmanabhan J, Chellappan SP. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene. 2002;21:8388-8396. [Cited in This Article: ] |
| 25. | Wang S, Nath N, Fusaro G, Chellappan S. Rb and prohibitin target distinct regions of E2F1 for repression and respond to different upstream signals. Mol Cell Biol. 1999;19:7447-7460. [Cited in This Article: ] |