Published online Sep 7, 2007. doi: 10.3748/wjg.v13.i33.4431
Revised: May 8, 2007
Accepted: May 12, 2007
Published online: September 7, 2007
Hepatitis C virus (HCV) infection is one of the major causes of chronic liver disease, including cirrhosis and liver cancer and is therefore, the most common indication for liver transplantation. Conventional antiviral drugs such as pegylated interferon-alpha, taken in combination with ribavirin, represent a milestone in the therapy of this disease. However, due to different viral and host factors, clinical success can be achieved only in approximately half of patients, making urgent the requirement of exploiting alternative approaches for HCV therapy. Fortunately, recent advances in the understanding of HCV viral replication and host cell interactions have opened new possibilities for therapeutic intervention. The most recent technologies, such as small interference RNA mediated gene-silencing, anti-sense oligonucleotides (ASO), or viral vector based gene delivery systems, have paved the way to develop novel therapeutic modalities for HCV. In this review, we outline the application of these technologies in the context of HCV therapy. In particular, we will focus on the newly defined role of cellular microRNA (miR-122) in viral replication and discuss its potential for HCV molecular therapy.
- Citation: Pan QW, Henry SD, Scholte BJ, Tilanus HW, Janssen HL, Laan LJVD. New therapeutic opportunities for Hepatitis C based on small RNA. World J Gastroenterol 2007; 13(33): 4431-4436
- URL: https://www.wjgnet.com/1007-9327/full/v13/i33/4431.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i33.4431
Hepatitis C virus (HCV), first identified in 1989, is a single-stranded positive-sense RNA flavivirus with 6 major genotypes and over 70 subtypes[1,2]. According to the estimation of the World Health Organization, approximately 170 million people, 3% of the world population, are HCV positive with 3 to 4 million de novo infections each year. Unfortunately, 55%-85% of those infected fail to clear the virus and progress to develop chronic infection. Over a period of 20 to 30 years cirrhosis develops in about 10% to 20% and hepatocellular carcinoma (HCC) develops in 1% to 7% of persons with chronic infection[3]. Currently, no safe and effective vaccine is available to prevent HCV infection. Conventional treatment, such as interferon taken alone or in combination with ribavirin, is only effective in part of the patients, but is often financially inaccessible for people in developing countries[4,5].
To explore the potential of new therapeutic strategies, it is critical to better understand the viral and host factors involved in virus cell entry, replication and virus-cell interaction. An apparent two-way dialogue exists in which the virus apparently takes advantage of the cells’ own signal transduction systems to facilitate virus entry and support replication[6]. Indeed, remarkable progress has been achieved in understanding the properties of the HCV genome and viral proteins. Contributions have come through several different sources, including vaccination of chimpanzees, structural studies, binding studies with recombinant envelope proteins, and the use of clinical isolates, HCV-like particles (HCV-LPs), HCV pseudotyped particles (HCVpp), and cell culture-derived HCV particles (HCVcc) in infectivity assays[7,8]. Cellular pathways or molecules involved in viral entry, such as CD81, scavenger receptor class B type I (SR-BI), LDL receptor, L-SIGN, DC-SIGN and asialoglycoprotein receptor (ASGPR) could be putative therapeutic targets[9-12].
New technologies, particularly RNA interference (RNAi) induced by small interfering RNA (siRNA), are gaining favour as effective therapeutic entities for HCV infections. RNAi works at a posttranscriptional level by degrading cognate mRNA. As HCV is a single-stranded RNA that functions as both a messenger RNA and a template for replication, it is a prime candidate for RNAi. Moreover, previous reports have shown that by blocking cellular determinants of viral entry and replication, such as CD81, HSP90, or p68, either by RNAi, antisense oligonucleotides or chemically engineered “antagomirs”, leads to significant reduction of viral invasion[13-15]. In this review, we outline the novel small RNA based technologies in designing therapeutic approaches for HCV treatment, according to the mechanism of viral entry, replication and virus-cell interaction. In particular, we will discuss emerging evidence that a liver-specific, small non-coding microRNA (miRNA) is involved in replication of HCV through a novel mechanism and outline its therapeutic potential.
HCV, contains a single-stranded RNA genome of about 9400 nucleotides in length, composed of a 5′ and 3′ non-coding region (NCR) with a single open reading frame encoding a polyprotein precursor of approximately 3000 amino acids that is cleaved into three structural (core, E1, E2) and seven non-structural (p7, NS2-NS5B) proteins[16,17].
Since the discovery of HCV, numerous studies have demonstrated its mechanism of cell entry, but it is still unclear how the virus penetrates cell membranes. In order to elucidate the infection pathway, it is first required to identify and understand both the putative viral and cell factors involved in this process. The viral envelope glycoproteins E1 and E2, cleaved from the polyprotein by the endoplasmic reticulum (ER)-resident host enzymes signal peptidase and signal peptide peptidase, have been widely regarded as the critical determinants for virus cell entry. To date, several models have been designed to investigate E1/E2 function. These include HCV-LPs expressing E1-E2 heterodimers instead of glycosylated individual E1 and E2[18-21], HCVpp consisting of unmodified HCV envelope glycoproteins E1 and E2 assembled onto retroviral or lentiviral core particles[22-26], vesicular stomatitis virus (VSV)/HCV pseudotypes expressing HCV E1 or E2 chimeric proteins containing transmembrane and cytoplasmic domains of the VSV G glycoprotein, or HCVcc neutralization assays with E1 or E2 antibody[27-30]. These models have shown that both envelope glycoproteins E1 and E2 are essential for host cell entry. The lack of either E1 or E2 significantly decreases HCV infection activity whereas deletion of the whole envelope protein coding sequence abolishes the particle infectivity. Additionally, several cell surface molecules have been identified using these models and are now considered as critical components in mediating HCV attachment and entry.
Similar to viral entry, HCV replication requires both viral and cellular factors. Although our current knowledge of the HCV life cycle is still mainly at the hypothetical level, several minimum viral components and host cell factors have been proposed. The HCV 5′ NCR, in particular the IRES sequence, plays an important function in ribosomal assembly and the NS3 to NS5B coding region are necessary for function of the replicase complex[31-35]. Found as interaction partners of NS5A and NS5B, human vesicle-associated membrane protein-associated proteins VAP-A and VAP-B were first identified from the host cell[36,37]. More recently, the geranylgeranylated protein FBL-2, the immunophilins cyclophilin B and FKBP8 have been identified as important host factors for HCV replication[38-40]. Furthermore, the host enzyme IMPDH, essential for the de novo synthesis of GTP nucleotides, may be involved in HCV replication as the IMPDH inhibitors ribavirin and mycophenolic acid suppresses replication[41,42]. Interestingly, the mammalian liver-specific miRNA (miR-122) has been recently defined to facilitate HCV replication, indicating that this small RNA may present a novel target for antiviral intervention[43].
miRNAs are approximately 22 nucleotide noncoding RNAs that can downregulate various gene products by inducing either cleavage or a reduction in the translational efficiency of the target mRNA[44,45]. In the last 5 years, over 3000 miRNAs have been identified in vertebrates, flies, worms, plants and even viruses. Most miRNAs have been shown to participate in essential biological processes, such as cell proliferation, apoptosis, differentiation and metabolism[46]. The 22 nucleotide mature miR-122, derived from a noncoding polyadenylated RNA transcript of the hcr gene, is a liver-specific developmental regulator. It can be detected as early as 12.5 d post-gestation and reach a plateau immediately before birth, then slowly increase up to 70% of the total miRNA population in adult liver[47-49]. miR-122 is the first identified host miRNA linked to HCV viral replication. A further novelty to these findings is the fact that miR-122 upregulates, rather than downregulates, viral RNA by interaction with the 5′ NCR of the viral RNA. Previous work had suggested that miRNA can only negatively regulate gene expression through targeting the 3′ NCR of mRNA.
Interestingly, Jopling et al[43] have observed that though both Huh7 and HepG2 cells are derived from human hepatocytes, HCV RNA can only replicate in Huh7 cells. This may link to the fact that Huh7 is miR-122 positive, while HepG2 is miR-122 negative. To determine if miR-122 is required to regulate HCV replication, they transfected antisense oligonucleotides into Huh7 liver cells to suppress miR-122 function. The results showed that the amount of viral RNA was reduced by about 80% when miR-122 was silenced, but it is still unclear whether it is simply a direct or indirect interaction through cellular factors. Thus, to further address this issue, two putative binding sites, located in each of the viral NCR, were tested as possible targets for miR-122. It was found that only the binding sequence located in the 5′ NCR was responsible for miR-122 targeting. This is notably very different from the common observation that miRNA target the 3′ NCR, leading to suppression or degradation of target mRNA. Recently, a study in mice has shown synthesized antisense single-stranded 23-nucleotide RNA molecules can effectively inhibit production of miR-122 in vivo[50]. Therefore, miR-122 seems a potential target for HCV treatment, although the mechanism for this new miRNA role is still very much unclear.
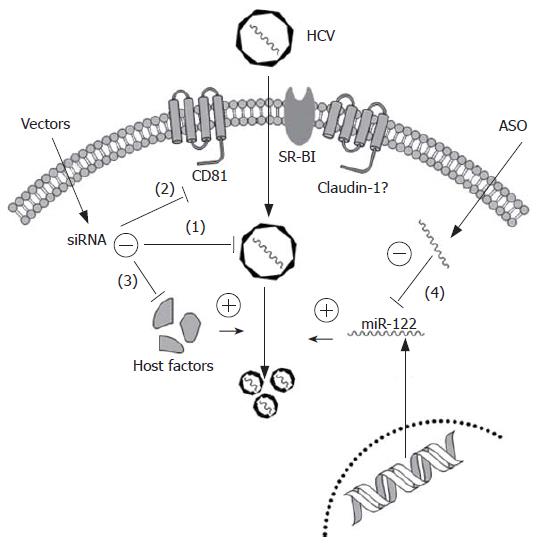
As current antiviral regimens have proven largely unsatisfactory, particularly for patients with genotype 1 infection, it is important to explore novel therapeutic strategies. Small interfering RNAs and antisense oligonucleotides (ASO) have emerged as efficient nuclei acid-based gene silencing tools to target highly conserved or functionally important regions within the HCV genome or essential host cell factors for entry or replication (Figure 1).
RNAi, induces gene silencing at a post-transcription level by double-stranded small interference RNA (siRNA) and represents an exciting new technology that could have applications in the treatment of viral diseases. Particularly, HCV could be an attractive target for RNAi therapy, as it is a RNA virus. The HCV genome is a positive single-stranded RNA that functions both as the viral messenger RNA and a template for RNA replication via a negative-strand intermediate. Instead of a 5' cap, the IRES, located at the 5' NCR, plays an essential role to bind eukaryotic ribosomal subunits and initiates the assembly of the translationally active 80S complex. Consequently, this sequence is more conserved than any other part of the viral genome, at least among the six known HCV genotypes[51,52]. Thus, IRES seems an ideal target for RNAi mediated anti-HCV therapy and several groups have demonstrated efficient inhibition of HCV replication by designing siRNAs toward this region[53-55]. In addition, RNAi directed against the viral core, NS3, NS4B, NS5A and NS5B regions can suppress HCV infection. McCaffrey et al[56] was the first to demonstrate feasibility of siRNA targeting HCV NS5B in vivo. By co-expression of an NS5B-luciferase fusion gene with an anti-NS5B siRNA expression plasmid they found a significant reduction of luciferase expression in the mouse liver indicating selective degradation by the NS5B siRNA. Additionally, several other groups have observed suppression of HCV replicon by siRNA-mediated targeting either NS5B or NS3 region[57-59].
Besides these viral elements, numerous host cellular factors, such as CD81, SR-BI, HSP90, p68 or USP18, could be typical targets for potentiating RNAi antiviral therapy. CD81, expressed in most human cells, is able to bind to HCV E2 protein and is, therefore, considered an essential receptor for HCV entry. Further investigation, by either ectopic expression of CD81 in Huh7-Lunet cells (low expression of CD81) or modulation of CD81 cell surface density in Huh-7.5 cells (high expression of CD81) by RNAi, revealed that density of cell surface-exposed CD81 is a key determinant for HCV entry into host cells[60]. SR-BI, primarily expressed in the liver and steroidogenic tissues, was identified as another potential HCV receptor based on coprecipitation with recombinant E2. A 90% down-regulation of SR-BI expression in Huh7 cells by RNAi caused a 30%-90% inhibition of HCVpp infection, depending on the HCV genotype[61,62]. However, either CD81 or SR-BI alone is not capable of virus binding indicating that at least one additional host protein, possibly the recently identified co-receptor, Claudin-1[63], is required for cell entry of enveloped virions via the CD81/SR-BI pathways.
Although using siRNA to target either viral or host factors could be considered effective tools to significantly block HCV infection and replication, an advanced method by knockdown both viral and cellular factors may further improve the therapeutic efficacy. Work by our group has shown that both entry and replication can be simultaneously targeted using shRNAs directed against two regions of the HCV RNA and one region of the host cell receptor, CD81. The triple shRNA expression vector was effective in concurrently reducing HCV replication, CD81 expression, and E2 binding, comparable to conventional single shRNA anti-HCV vectors[64].
Antisense oligonucleotides represent an alternative gene-silencing tool that can be employed as HCV therapy. ASO-based inhibition of HCV has been demonstrated extensively in the past[65-71]. Currently, ASO is the most promising method to block the function of miRNA, such as miR-122. For instance, a 2′-O-methylated RNA oligonucleotide with exact complementarity to miR-122 was introduced to inactivate its function in Huh7 cells, in order to determine the relationship between miR-122 and HCV replication. Subsequently, Krutzfeldt et al[50] developed a pharmacological approach for silencing miRNA in vivo, by chemically modified, cholesterol-conjugated single-stranded RNA analogues to complementarily target miR-122. By injection of these ‘antagomirs’ into the tail veins of mice, efficient and specific suppression of endogenous miR-122 was observed. Hence, designing ASO based molecular medicines would provide new agents for human major diseases, because upregulation of certain miRNAs linked to a set of diseases such as cancer, diabetes or HCV.
Obviously, RNAi or ASO technologies could be regarded as potentially effective novel modalities for anti-HCV treatment. Nevertheless, the success depends on developing effective delivery systems, to target therapy to the liver. Regarding to treat a liver-hosted and long-term persistent hepatitis virus, an ideal vector would be able to transfer genetic material efficiently and specifically into the target cells/tissues, resulting in high level, properly regulated and prolonged expression, without toxic and immunogenic side effects. Since viruses have many advantages as transgenic vehicles, we will discuss two of the most promising delivery systems: lentiviral and adeno-associated viral (AAV) vectors.
Lentiviral vectors, are mainly based on human immunodeficiency virus type 1 (HIV-1) and have been shown to effectively transduce liver, muscle, and hematopoietic cells. These vectors integrate their payloads into the host genome ensuring transmission to progeny cells[72]. Although lentiviral-mediated short hairpin RNA (shRNA, precursor of siRNA) delivery has been widely developed for therapeutic application, there are few reports referring to HCV treatment[57,64]. There are currently some limitations for the use of lentiviral vectors: (1) production efficiency limits in vivo transfection; (2) possibility of insertional mutagenesis or generation of wild-type virus leading to safety considerations. To circumvent these drawbacks the following strategies may be required to achieve further improvement: firstly, newer generations, such as the gutted third generation, relatively high titers of VSV-G pseudotyped HIV-1 vectors, other types such as HIV-2 and simian immunodeficiency virus (SIV) vectors, or even immunodeficiency viruses derived from nonprimates, including felines and equines, are also being developed to overcome conventional problems[73-76].
Analogically, with the superiority of low pathogenicity and long-term gene expression, AAV could be another ideal viral vector for siRNA delivery, although no reference of AAV-mediated anti-HCV RNAi therapy has been reported so far. Particularly AAV serotype 8, a new member of the AAV family isolated from rhesus monkeys, is an attractive candidate for hepatic-directed shRNA transfer because of 10- to 100-fold increased transduction efficiency in mouse liver models, compared with the previous AAV2 based vectors[77]. Since derived from nonhuman primate, AAV8 is less prone to recognition by prevailing antibodies that generate side immunological effects in human[78]. Moreover, the safety and transgenic delivery efficacy could be further improved by conjugating other strategies, such as utilizing liver-specific promoters, hybridization of AAV8 with other serotypes, or modification of viral capsids.
Furthermore, since miRNA context based siRNA cassette (second-generation shRNA) can be driven by a regulated polII promoter instead of conventional polIII promoters[79], liver-targeted expression of shRNA could be achieved by employing a liver-specific polII promoter in viral delivery system.
The treatment of HCV remains a challenge that requires further elucidating the process of viral life cycle and developing novel therapeutic approaches. In fact, recent progress has provided the possibilities of identifying novel antiviral targets and designing new therapeutic strategies. According to the previous description, miR-122 is one of the most emergent targets for HCV therapy that is commonly abundant in human livers and thus promotes viral replication. Therefore, downregulation of miR-122 by antisense based ‘antagomirs’ or oligonucleotides significantly suppressed viral replication. However, before such a method can be applied in the clinic, the role of miR-122 in maintaining normal hepatic function must be further investigated. Krutzfeldt et al[50] have demonstrated that silencing of miR-122 by ‘antagomirs’ do not show any apparent toxicity to mice, but the more recent study has shown that miR-122 is downregulated in the rodent and human hepatocellular carcinomas (HCC). Using the animal model of diet-induced hepatocarcinogenesis, Kutay et al[80] have observed that the reduced expression of miR-122 probably occurs between 36 and 54 wk when neoplastic transformation occurs. These findings suggest that the downregulation of miR-122 might be associated with hepatocarcinogenesis and, therefore, further investigation into the function of miR-122 is required before therapeutic application can be commenced. In conclusion, the recent progress of understanding the viral life cycle and identification of novel targets, in combination with the newly developed ASO and RNAi technology, may pave the way for new anti-HCV therapy.
We would like to thank Dr. Herold J Metselaar for his general support. S.D.H. is supported by a “Translational research” grant of the Erasmus MC, Rotterdam.
S- Editor Liu Y L- Editor Rippe RA E- Editor Liu Y
| 1. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4996] [Cited by in F6Publishing: 4592] [Article Influence: 131.2] [Reference Citation Analysis (0)] |
| 2. | Simmonds P, Alberti A, Alter HJ, Bonino F, Bradley DW, Brechot C, Brouwer JT, Chan SW, Chayama K, Chen DS. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology. 1994;19:1321-1324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 619] [Cited by in F6Publishing: 530] [Article Influence: 17.7] [Reference Citation Analysis (1)] |
| 3. | Sarbah SA, Younossi ZM. Hepatitis C: an update on the silent epidemic. J Clin Gastroenterol. 2000;30:125-143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 95] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 4. | Hoofnagle JH, di Bisceglie AM. The treatment of chronic viral hepatitis. N Engl J Med. 1997;336:347-356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 715] [Cited by in F6Publishing: 723] [Article Influence: 26.8] [Reference Citation Analysis (1)] |
| 5. | McHutchison JG, Patel K. Future therapy of hepatitis C. Hepatology. 2002;36:S245-S252. [PubMed] [Cited in This Article: ] |
| 6. | Smith AE, Helenius A. How viruses enter animal cells. Science. 2004;304:237-242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 561] [Cited by in F6Publishing: 548] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 7. | Bukh J, Forns X, Emerson SU, Purcell RH. Studies of hepatitis C virus in chimpanzees and their importance for vaccine development. Intervirology. 2001;44:132-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 44] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Cocquerel L, Voisset C, Dubuisson J. Hepatitis C virus entry: potential receptors and their biological functions. J Gen Virol. 2006;87:1075-1084. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 141] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 9. | Gardner JP, Durso RJ, Arrigale RR, Donovan GP, Maddon PJ, Dragic T, Olson WC. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc Natl Acad Sci USA. 2003;100:4498-4503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 230] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 10. | Lozach PY, Lortat-Jacob H, de Lacroix de Lavalette A, Staropoli I, Foung S, Amara A, Houles C, Fieschi F, Schwartz O, Virelizier JL. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J Biol Chem. 2003;278:20358-20366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 281] [Cited by in F6Publishing: 285] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 11. | Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G. Binding of hepatitis C virus to CD81. Science. 1998;282:938-941. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1572] [Cited by in F6Publishing: 1521] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 12. | Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017-5025. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 888] [Cited by in F6Publishing: 868] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 13. | Goh PY, Tan YJ, Lim SP, Tan YH, Lim SG, Fuller-Pace F, Hong W. Cellular RNA helicase p68 relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication. J Virol. 2004;78:5288-5298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 92] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 14. | Keck ZY, Xia J, Cai Z, Li TK, Owsianka AM, Patel AH, Luo G, Foung SK. Immunogenic and functional organization of hepatitis C virus (HCV) glycoprotein E2 on infectious HCV virions. J Virol. 2007;81:1043-1047. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 77] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 15. | Nakagawa S, Umehara T, Matsuda C, Kuge S, Sudoh M, Kohara M. Hsp90 inhibitors suppress HCV replication in replicon cells and humanized liver mice. Biochem Biophys Res Commun. 2007;353:882-888. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 74] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 16. | Giannini C, Bréchot C. Hepatitis C virus biology. Cell Death Differ. 2003;10 Suppl 1:S27-S38. [PubMed] [Cited in This Article: ] |
| 17. | Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 458] [Cited by in F6Publishing: 490] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 18. | Wellnitz S, Klumpp B, Barth H, Ito S, Depla E, Dubuisson J, Blum HE, Baumert TF. Binding of hepatitis C virus-like particles derived from infectious clone H77C to defined human cell lines. J Virol. 2002;76:1181-1193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 81] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Barth H, Ulsenheimer A, Pape GR, Diepolder HM, Hoffmann M, Neumann-Haefelin C, Thimme R, Henneke P, Klein R, Paranhos-Baccalà G. Uptake and presentation of hepatitis C virus-like particles by human dendritic cells. Blood. 2005;105:3605-3614. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 73] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 20. | Steinmann D, Barth H, Gissler B, Schürmann P, Adah MI, Gerlach JT, Pape GR, Depla E, Jacobs D, Maertens G. Inhibition of hepatitis C virus-like particle binding to target cells by antiviral antibodies in acute and chronic hepatitis C. J Virol. 2004;78:9030-9040. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | Triyatni M, Saunier B, Maruvada P, Davis AR, Ulianich L, Heller T, Patel A, Kohn LD, Liang TJ. Interaction of hepatitis C virus-like particles and cells: a model system for studying viral binding and entry. J Virol. 2002;76:9335-9344. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 102] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 22. | Bartosch B, Bukh J, Meunier JC, Granier C, Engle RE, Blackwelder WC, Emerson SU, Cosset FL, Purcell RH. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc Natl Acad Sci USA. 2003;100:14199-14204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 241] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 23. | Lavillette D, Morice Y, Germanidis G, Donot P, Soulier A, Pagkalos E, Sakellariou G, Intrator L, Bartosch B, Pawlotsky JM. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J Virol. 2005;79:6023-6034. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 209] [Cited by in F6Publishing: 209] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 24. | Logvinoff C, Major ME, Oldach D, Heyward S, Talal A, Balfe P, Feinstone SM, Alter H, Rice CM, McKeating JA. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc Natl Acad Sci USA. 2004;101:10149-10154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 333] [Cited by in F6Publishing: 324] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 25. | Meunier JC, Engle RE, Faulk K, Zhao M, Bartosch B, Alter H, Emerson SU, Cosset FL, Purcell RH, Bukh J. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc Natl Acad Sci USA. 2005;102:4560-4565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 207] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 26. | Schofield DJ, Bartosch B, Shimizu YK, Allander T, Alter HJ, Emerson SU, Cosset FL, Purcell RH. Human monoclonal antibodies that react with the E2 glycoprotein of hepatitis C virus and possess neutralizing activity. Hepatology. 2005;42:1055-1062. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 56] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 27. | Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972-1974. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1154] [Cited by in F6Publishing: 1139] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 28. | Buonocore L, Blight KJ, Rice CM, Rose JK. Characterization of vesicular stomatitis virus recombinants that express and incorporate high levels of hepatitis C virus glycoproteins. J Virol. 2002;76:6865-6872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 76] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 29. | Dreux M, Pietschmann T, Granier C, Voisset C, Ricard-Blum S, Mangeot PE, Keck Z, Foung S, Vu-Dac N, Dubuisson J. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J Biol Chem. 2006;281:18285-18295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 170] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 30. | Lagging LM, Meyer K, Owens RJ, Ray R. Functional role of hepatitis C virus chimeric glycoproteins in the infectivity of pseudotyped virus. J Virol. 1998;72:3539-3546. [PubMed] [Cited in This Article: ] |
| 31. | Brass V, Bieck E, Montserret R, Wölk B, Hellings JA, Blum HE, Penin F, Moradpour D. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J Biol Chem. 2002;277:8130-8139. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 281] [Cited by in F6Publishing: 295] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 32. | Egger D, Wölk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974-5984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 639] [Cited by in F6Publishing: 617] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 33. | Penin F, Brass V, Appel N, Ramboarina S, Montserret R, Ficheux D, Blum HE, Bartenschlager R, Moradpour D. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J Biol Chem. 2004;279:40835-40843. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 221] [Cited by in F6Publishing: 233] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 34. | Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513-1515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 231] [Cited by in F6Publishing: 236] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 35. | Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959-1962. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 393] [Cited by in F6Publishing: 416] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 36. | Evans MJ, Rice CM, Goff SP. Genetic interactions between hepatitis C virus replicons. J Virol. 2004;78:12085-12089. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 37. | Hamamoto I, Nishimura Y, Okamoto T, Aizaki H, Liu M, Mori Y, Abe T, Suzuki T, Lai MM, Miyamura T. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J Virol. 2005;79:13473-13482. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 162] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 38. | Okamoto T, Nishimura Y, Ichimura T, Suzuki K, Miyamura T, Suzuki T, Moriishi K, Matsuura Y. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 2006;25:5015-5025. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 202] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 39. | Wang C, Gale M, Keller BC, Huang H, Brown MS, Goldstein JL, Ye J. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell. 2005;18:425-434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 217] [Cited by in F6Publishing: 226] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 40. | Watashi K, Ishii N, Hijikata M, Inoue D, Murata T, Miyanari Y, Shimotohno K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol Cell. 2005;19:111-122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 351] [Cited by in F6Publishing: 366] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 41. | Dixit NM, Perelson AS. The metabolism, pharmacokinetics and mechanisms of antiviral activity of ribavirin against hepatitis C virus. Cell Mol Life Sci. 2006;63:832-842. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 94] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 42. | Henry SD, Metselaar HJ, Lonsdale RC, Kok A, Haagmans BL, Tilanus HW, van der Laan LJ. Mycophenolic acid inhibits hepatitis C virus replication and acts in synergy with cyclosporin A and interferon-alpha. Gastroenterology. 2006;131:1452-1462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 92] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 43. | Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577-1581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1993] [Cited by in F6Publishing: 1929] [Article Influence: 101.5] [Reference Citation Analysis (0)] |
| 44. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25833] [Cited by in F6Publishing: 26778] [Article Influence: 1338.9] [Reference Citation Analysis (0)] |
| 45. | He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522-531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4964] [Cited by in F6Publishing: 5118] [Article Influence: 255.9] [Reference Citation Analysis (0)] |
| 46. | Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev. 2005;15:410-415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 268] [Cited by in F6Publishing: 266] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 47. | Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 607] [Cited by in F6Publishing: 632] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 48. | Etiemble J, Möröy T, Jacquemin E, Tiollais P, Buendia MA. Fused transcripts of c-myc and a new cellular locus, hcr in a primary liver tumor. Oncogene. 1989;4:51-57. [PubMed] [Cited in This Article: ] |
| 49. | Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735-739. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2502] [Cited by in F6Publishing: 2504] [Article Influence: 113.8] [Reference Citation Analysis (0)] |
| 50. | Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685-689. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3005] [Cited by in F6Publishing: 2983] [Article Influence: 157.0] [Reference Citation Analysis (0)] |
| 51. | Krönke J, Kittler R, Buchholz F, Windisch MP, Pietschmann T, Bartenschlager R, Frese M. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J Virol. 2004;78:3436-3446. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 133] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 52. | Prabhu R, Garry RF, Dash S. Small interfering RNA targeted to stem-loop II of the 5' untranslated region effectively inhibits expression of six HCV genotypes. Virol J. 2006;3:100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 53. | Hamazaki H, Ujino S, Abe E, Miyano-Kurosaki N, Shimotohno K, Takaku H. RNAi expression mediated inhibition of HCV replication. Nucleic Acids Symp Ser (Oxf). 2004;307-308. [PubMed] [Cited in This Article: ] |
| 54. | Ilves H, Kaspar RL, Wang Q, Seyhan AA, Vlassov AV, Contag CH, Leake D, Johnston BH. Inhibition of hepatitis C IRES-mediated gene expression by small hairpin RNAs in human hepatocytes and mice. Ann N Y Acad Sci. 2006;1082:52-55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 55. | Kanda T, Steele R, Ray R, Ray RB. Small interfering RNA targeted to hepatitis C virus 5' nontranslated region exerts potent antiviral effect. J Virol. 2007;81:669-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 68] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 56. | McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38-39. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 798] [Cited by in F6Publishing: 756] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 57. | Takigawa Y, Nagano-Fujii M, Deng L, Hidajat R, Tanaka M, Mizuta H, Hotta H. Suppression of hepatitis C virus replicon by RNA interference directed against the NS3 and NS5B regions of the viral genome. Microbiol Immunol. 2004;48:591-598. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 58. | Wilson JA, Richardson CD. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J Virol. 2005;79:7050-7058. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 153] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 59. | Zhang J, Yamada O, Sakamoto T, Yoshida H, Araki H, Murata T, Shimotohno K. Inhibition of hepatitis C virus replication by pol III-directed overexpression of RNA decoys corresponding to stem-loop structures in the NS5B coding region. Virology. 2005;342:276-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 60. | Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol. 2007;81:588-598. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 179] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 61. | Bartosch B, Verney G, Dreux M, Donot P, Morice Y, Penin F, Pawlotsky JM, Lavillette D, Cosset FL. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J Virol. 2005;79:8217-8229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 227] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 62. | Lavillette D, Tarr AW, Voisset C, Donot P, Bartosch B, Bain C, Patel AH, Dubuisson J, Ball JK, Cosset FL. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology. 2005;41:265-274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 209] [Cited by in F6Publishing: 219] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 63. | Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801-805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 930] [Cited by in F6Publishing: 915] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 64. | Henry SD, van der Wegen P, Metselaar HJ, Tilanus HW, Scholte BJ, van der Laan LJ. Simultaneous targeting of HCV replication and viral binding with a single lentiviral vector containing multiple RNA interference expression cassettes. Mol Ther. 2006;14:485-493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 85] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 65. | el-Awady MK, el-Din NG, el-Garf WT, Youssef SS, Omran MH, el-Abd J, Goueli SA. Antisense oligonucleotide inhibition of hepatitis C virus genotype 4 replication in HepG2 cells. Cancer Cell Int. 2006;6:18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 66. | Hanecak R, Brown-Driver V, Fox MC, Azad RF, Furusako S, Nozaki C, Ford C, Sasmor H, Anderson KP. Antisense oligonucleotide inhibition of hepatitis C virus gene expression in transformed hepatocytes. J Virol. 1996;70:5203-5212. [PubMed] [Cited in This Article: ] |
| 67. | Heintges T, Encke J, zu Putlitz J, Wands JR. Inhibition of hepatitis C virus NS3 function by antisense oligodeoxynucleotides and protease inhibitor. J Med Virol. 2001;65:671-680. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 68. | McCaffrey AP, Meuse L, Karimi M, Contag CH, Kay MA. A potent and specific morpholino antisense inhibitor of hepatitis C translation in mice. Hepatology. 2003;38:503-508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 69. | McHutchison JG, Patel K, Pockros P, Nyberg L, Pianko S, Yu RZ, Dorr FA, Kwoh TJ. A phase I trial of an antisense inhibitor of hepatitis C virus (ISIS 14803), administered to chronic hepatitis C patients. J Hepatol. 2006;44:88-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 70. | Wakita T, Wands JR. Specific inhibition of hepatitis C virus expression by antisense oligodeoxynucleotides. In vitro model for selection of target sequence. J Biol Chem. 1994;269:14205-14210. [PubMed] [Cited in This Article: ] |
| 71. | Zhang H, Hanecak R, Brown-Driver V, Azad R, Conklin B, Fox MC, Anderson KP. Antisense oligonucleotide inhibition of hepatitis C virus (HCV) gene expression in livers of mice infected with an HCV-vaccinia virus recombinant. Antimicrob Agents Chemother. 1999;43:347-353. [PubMed] [Cited in This Article: ] |
| 72. | Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263-267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3710] [Cited by in F6Publishing: 3600] [Article Influence: 128.6] [Reference Citation Analysis (0)] |
| 73. | Johnston JC, Gasmi M, Lim LE, Elder JH, Yee JK, Jolly DJ, Campbell KP, Davidson BL, Sauter SL. Minimum requirements for efficient transduction of dividing and nondividing cells by feline immunodeficiency virus vectors. J Virol. 1999;73:4991-5000. [PubMed] [Cited in This Article: ] |
| 74. | Poeschla E, Gilbert J, Li X, Huang S, Ho A, Wong-Staal F. Identification of a human immunodeficiency virus type 2 (HIV-2) encapsidation determinant and transduction of nondividing human cells by HIV-2-based lentivirus vectors. J Virol. 1998;72:6527-6536. [PubMed] [Cited in This Article: ] |
| 75. | Poeschla EM, Wong-Staal F, Looney DJ. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat Med. 1998;4:354-357. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 323] [Cited by in F6Publishing: 322] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 76. | Rizvi TA, Panganiban AT. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J Virol. 1993;67:2681-2688. [PubMed] [Cited in This Article: ] |
| 77. | Nakai H, Fuess S, Storm TA, Muramatsu S, Nara Y, Kay MA. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J Virol. 2005;79:214-224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 245] [Cited by in F6Publishing: 255] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 78. | Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. Clades of Adeno-associated viruses are widely disseminated in human tissues. J Virol. 2004;78:6381-6388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 717] [Cited by in F6Publishing: 765] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 79. | Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281-1288. [PubMed] [Cited in This Article: ] |
| 80. | Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671-678. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 450] [Cited by in F6Publishing: 492] [Article Influence: 28.9] [Reference Citation Analysis (0)] |