Published online Sep 7, 2006. doi: 10.3748/wjg.v12.i33.5265
Revised: May 28, 2006
Accepted: July 7, 2006
Published online: September 7, 2006
Alcohol consumption causes cellular injury. Recent developments indicate that ethanol induces epigenetic alterations, particularly acetylation, methylation of histones, and hypo- and hypermethylation of DNA. This has opened up a new area of interest in ethanol research and is providing novel insight into actions of ethanol at the nucleosomal level in relation to gene expression and patho-physiological consequences. The epigenetic effects are mainly attributable to ethanol metabolic stress (Emess), generated by the oxidative and non-oxidative metabolism of ethanol, and dysregulation of methionine metabolism. Epigenetic changes are important in ethanol-induced hepatic steatosis, fibrosis, carcinoma and gastrointestinal injury. This editorial highlights these new advances and its future potential.
- Citation: Shukla SD, Aroor AR. Epigenetic effects of ethanol on liver and gastrointestinal injury. World J Gastroenterol 2006; 12(33): 5265-5271
- URL: https://www.wjgnet.com/1007-9327/full/v12/i33/5265.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i33.5265
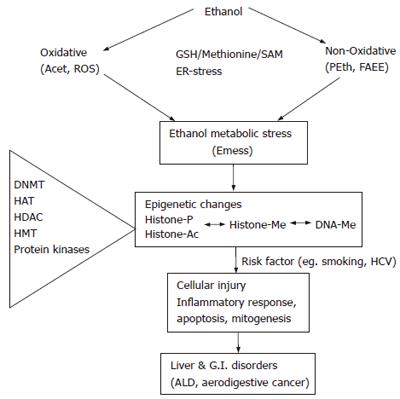
Ethanol actions are diverse and fascinatingly complex. Chronic ethanol causes injury to almost all organ systems including liver and gastrointestine (GI)[1] and has serious medical and public health implications[2]. Alcohol increases the risk for hepatocellular carcinoma (HCC) and colon cancer. Although these effects of ethanol are now widely known, our knowledge on the mechanisms of actions of ethanol at the subcellular and molecular levels is poor. Therapeutic tools to control or reverse the ethanol-induced cellular damages, such as alcoholic liver injury, are also lacking. In addition to its direct actions, ethanol-induced effects are also mediated by oxidative [e.g. acetaldehyde, reactive oxygen species (ROS)] and non-oxidative [e.g. phosphatidylethanol (PEth), fatty acid ethyl ester (FAEE)] metabolites/products and impairment in the methylation process. It is the combination of these metabolic stress pathways, termed as “ethanol metabolic stress” (Emess), which contributes to the epigenetic effects of ethanol (Figure 1).
The question of how a single cell can differentiate into many different cell types in a multicellular organism has long led to the hypothesis that additional information that regulates genomic functions must exist beyond the level of the genetic code. This concept led to the introduction of the term ‘epigenetics’ in the 1940’s, a term that has now evolved to mean heritable changes in gene expression that do not involve changes in DNA sequence[3-5]. Interestingly, these epigenetic changes are heritable and normally stably maintained. They are also reversible. The molecular basis of epigenetics has largely focused on mechanisms such as DNA methylation and histone modification. In fact, emerging evidence indicates that both mechanisms act in concert to provide stable and heritable silencing.
DNA methylation specifically occurs at the C5 position of cytosine residues that are associated with CpG dinucleotides. Eighty percent of all CpG dinucleotides in the mammalian genome are methylated. The remaining unmethylated CpG residues are mostly located in the promoter regions of constitutively active genes and are referred to as CpG islands. Methylation of DNA is known to modulate transcriptional repression, genomic imprinting and modulation of chromatin structure[4,5].
Global hypomethylation involves mainly repetitive sequences but hypomethylation of coding regions may also occur[6]. Hypermethylation of normally unmethylated genes can result in silencing of tumor suppressor genes. Stepwise distinct methylation events are likely to be the features of the sequence from hepatitis to HCC and may contribute to the process of hepatic carcinogenesis[7,8]. Regional hypermethylation and global hypomethylation are also well recognized in gastrointestinal cancer[9-12].
Only a few studies have addressed regional methylation of DNA in relation to alcohol and cancer. Alcohol either alone or in combination with tobacco has been shown to be an important risk factor for oral cancer[13,14]. Promoter hypermethylation of p16INK4a, p14ARF, RB1, p21Waf1, p27Kip1, PTEN, p73, O6-methyl guanine DNA methyl transferase (O6-MGMT), and GST-P genes has been examined in relation to smoking and alcohol use. Overall, gene methylation can be detectable in 46.9% of samples and is closely correlated with tobacco use and/or alcohol consumption[15]. The relative risk of alcohol consumption for the development of esophageal cancer is also very high[16] and alcohol potentiates chemical carcinogenesis of the esophagus induced by nitrosomethylbenzylamine[17]. Alcohol consumption has also been shown to be a risk factor for head and neck cancers that usually originates from the aerodigestive tract. Interestingly, p15 promoter hypermethylation has been observed in the healthy individuals who are smokers and/or alcohol consumers[18], suggesting that hypermethylation plays a significant role in progression of cancer. Although alcohol consumption is not a significant risk factor for gastric carcinoma compared to oral or esophageal cancer, both smoking and alcohol consumption are associated with a higher risk of gastric cancer with hypermethylation of the hMLH1 gene promoter. Hypermethylation of the hMLH1 gene promoter is inversely correlated with mutation of the p53 gene[19]. In a recent study, promoter hypermethylation of APC, p14 (ARF), p16 (INK4A), hMLH1, O6-MGMT, and RASSF1A was observed in colorectal cancer (CRC). For each of the tested genes, the prevalence of promoter hypermethylation is higher in CRCs derived from patients with low folate/high alcohol intake when compared with CRCs from patients with high folate/low alcohol intake[20].
Although methylation changes have been described as stable for aging and carcinoma, recent studies have shown that epigenetic alterations are also dynamic as observed in inflammatory responses and tissue injury[21]. Altered DNA methylation occurs after alcohol consumption during initial periods of alcohol abuse. Global hypomethylation of DNA in liver after long term ethanol exposure has been reported[22] but hypermethylation of DNA from peripheral blood cells after ethanol consumption has also been reported in human subjects with alcohol dependence[23]. Regional hypomethylation of the c-myc gene occurs in liver after long term consumption of alcohol[22]. Another study showed that chronic alcohol consumption produces global genomic DNA hypomethylation in the colonic mucosa[24].
Four known or putative mammalian DNA methyl transferases have been identified thus far: DNMT1, DNMT2, DNMT3a and DNMT3b. In contrast to types 1 and 3, the function of type 2 DNMT remains less clear. Mammalian DNMT2 does not methylate CGs. Non-CG cytosine methylation is also reported[25]. Increased expression of DNA methyl transferases occurs in hepatocellular carcinoma[26] and gastrointestinal cancer but this increased expression is associated with both hypomethylation and hypermethylation of DNA[27-29]. Decreased DNA methylation with a concomitant decrease in DNA methyl transferase activity after ethanol exposure of pregnant rats has been reported in fetal tissues[30]. Decreased activity of methyl transferase has been reported in peripheral blood cells from alcoholics but with a concomitant increase in DNA methylation[31]. This raises the possibility of regional methylations in a gene specific manner.
Chromatin is the entire DNA-protein complex packaged into chromosomes. It exists as a highly ordered structure and is composed of repeated nucleosome subunits. Each nucleosome contains a core of histone around which DNA is wrapped. Eukaryotes have five major classes of histones: H1, H2A, H2B, H3, and H4. Histones were once thought as static, non-participating structural elements; and now considered integral and dynamic components in the machinery responsible for regulating gene transcription[32]. The core histones (e.g. H3) have a similar structure with a basic N-terminal domain, a globular domain and a C-terminal tail. Modifications of histones can occur by mechanisms involving acetylation, phosphorylation, methylation, ubiquitination, sumoylation and ADP-ribosylation, etc. Some of these post-translational modifications affect packaging of genes, increase accessibility of transcription factors to DNA templates and initiate transcriptional processes[32,33]. Such modifications can serve as ‘co-activators’ (e.g. acetylation, methylation) or ‘co-repressors’ (e.g., deacetylation) or ‘gene silencers’ (e.g. methylation). In histone H3 from most species, the main acetylation sites include lysines 9, 14, 18 and 23. A steady state balance between two key enzymes, histone acetyl transferase (HAT) and histone deacetylase (HDAC), is crucial in this process. Various HATs[34] and HDACs[35] have been identified (about 15 types of HATs and 10 types of HDACs). H3 acetylation at lysine 9 or at lysine 14 plays a role in chromatin assembly[32], gene expression[36,37] and apoptosis[38]. Histones, particularly H3 and H4, are methylated at a number of lysines (Lys) and arginine residues. The major sites of lysine-methylation identified are: Lys4, Lys9, Lys27, Lys36, and Lys79 on H3 and Lys20 on H4. In addition, lysine residues can be methylated in the form of mono-, di- or trimethylation and this differential methylation provides further functional diversity to each site of Lys methylation[4,39]. Emerging evidence suggests that DNA and histone methylation likely have a cyclical and mutually reinforcing relationship, and both are required for stable and long-term epigenetic silencing[4,39,40]. Lysine 9 is also interesting in that this site can be either methylated (gene silencing) or acetylated (gene activation). Histone H3 can also be phosphorylated at ser-10 and ser-28 by cellular protein kinases[4,32,39]. The precise pattern of histone modification has been suggested to mediate biologically diverse effects and proposed as the ‘histone code’ hypothesis[32]. The relationship among Emess, histone modification, DNA methylation and changes in the expression level of genes is an emerging topic of investigation.
Initial studies with primary cultures of rat hepatocytes have established important characteristics of ethanol-induced histone acetylations. Ethanol causes a dose- and time-[41] dependent selective acetylation of histone H3 at Lys9 (H3AcK9). Other H3 lysine residues i.e. Lys14, Lys18 and Lys23 are not acetylated under these conditions. Trichostatin A, a reversible HDAC inhibitor, shows an increase in H3 acetylation. These increases in acetylation are not due to the increased expression of H3 protein as their levels do not change. It is also not due to the simple physical effect of ethanol since it requires more than 4 h of ethanol exposure to elicit H3 acetylation. The acetylation is reversible when ethanol is withdrawn after 24 h of treatment.
Ethanol causes activation of p42/44 MAPK, p38 MAPK and JNK in hepatocytes, while inhibition of p42/44 MAPK and JNK results in inhibition of ethanol-induced acetylation[42,43]. These results indicate that MAPK signaling plays a role in ethanol-induced epigenetic effects.
Ethanol acutely affects histone acetylation in vivo. Intragastric administration of ethanol increases 2-3 fold compared to the level of acetylated H3-Lys9 in the liver after 12 h, but has no effect on Lys14, Lys18 and Lys23. Further analysis indicates that the increased acetylation is tissue specific as it is noted in liver, lung and spleen but not in tissues from the brain, heart, kidney, muscle, vessels, stomach and intestine. Thus ethanol-induced histone H3 acetylation appears to be organ specific[44]. In rat liver stellate cells, ethanol increases H3 Lys 9 acetylation[45] but its significance remains to be determined.
Ethanol also affects histone H3 methylations in an interesting manner. The influence of ethanol on histone H3 Lys9 and Lys4 methylations in primary cultures of rat hepatocytes is determined using site specific antibodies. Western blot analysis using methylated forms of Lys4 and Lys9 histone H3 antibodies can show dramatically opposing changes in the methylated forms. The Lys9 methylation decreases but Lys4 methylation increases in hepatocytes. These results indicate that, like H3 acetylation, histone methylation is also sensitive to ethanol. A longer incubation with ethanol for 72 h does not change this methylation, indicating that ethanol-induced methylation produces a longer effect than that observed for acetylation which declines after 24 h (Bhadra, U and Shukla SD, Unpublished). Thus modifications in H3 methylation are likely to be coupled to hyperacetylation and orchestrate the fine tuning of the chromatin status in hepatocytes exposed to ethanol.
In hepatocytes exposed to ethanol, chromatin immunoprecipitation (CHIP) assays demonstrate the association of the acetylated H3-Lys9 with the alcohol dehydrogenase I (ADH 1) DNA domain in the nuclear chromatin[43]. These data argue that ethanol-elicited epigenetic changes cause an increased association between acetylated H3 and specific genes, a process which favors transcription[43]. It should be noted that circular dichroism spectrophotometry has shown altered chromatin confirmation in alcoholic rat liver, and this relaxed state of chromatin can promote transcription[46]. Thus ethanol modulates histone/chromatin to influence transcriptional activation. Further relevance of such epigenetic changes to the expression of genes involved in ethanol-induced tissue injury therefore merits investigation.
Although structural alterations in genes contributing to HCC are evident in transformed hepatocytes, initiation of hepatocarcinogenesis takes place during the early stages of liver insult and is associated with epigenetic alterations[7]. The progression of cell injury to carcinoma occurs due to triggering of ‘some’ molecular switches caused by a ‘second hit’, e.g. hepatitis C virus infection or other agents. Treatment of hepatocytes with ethanol causes apoptosis whereas alcohol enhances hepatic DNA synthesis in embryonic or transformed hepatocytes, through potentiation of G-protein mediated ras/MAPK signaling. This underscores the importance of normal versus embryonic or transformed hepatocytes contributing to the opposing effects of ethanol[47].
In this context, upregulation of ras signaling[48,49] concomitant with down regulation of p53-dependent apoptotic pathway[50-52] is seen in most cancers. Hyperme-thylation of apoptosis-related genes in ras transformed cells[53] and hypermethylation of genes implicated in apoptosis in HCC associated with alcohol consumption, viral infection and aflatoxin contamination have been reported[54]. Additionally, ras itself is subjected to epigenetic alteration by DNA methylation.
Hypomethylation of ras has been demonstrated in gastritis and gastric carcinoma[49]. Ethanol induces ras activation in gastric epithelial cells[55] and chronic alcoholic liver injury is associated with upregulation of ras activity[56]. C-myc, which regulates both apoptosis and proliferation, is overexpressed in HCC and cooperates with ras in the development of carcinoma[57]. Ethanol also causes an increased expression of c-myc, which is associated with hypomethylation of the c-myc gene[22].
p53 is also a modulator of histone acetylation and methylation[58,59]. Hyperacetylation of H3K9 with concomitant loss of dimethyl-H3K9 and increased methylation of H3K4 is seen with delayed suppression of hepatic alpha fetoprotein (AFP, a marker of embryonic phenotype) in p53-null mice[60]. There is loss of p53 function by its hypermethylation in hepatocellular carcinoma[61] and p53 mutation is common in gastrointestinal carcinoma[62]. Apoptosis in chronic alcoholic liver injury is associated with p53 accumulation[63]. In support of this, p53 null mice fed with ethanol exhibit suppression of apoptosis and increased proliferation of hepatocyets[64]. The preceding observations strongly indicate that ras and p53 as switch targets play a role in ethanol-induced epigenetic mechanisms.
Actions of ethanol are unique in that, ethanol or its metabolites have their own effects and can also sensitize (or desensitize) responses to other agonists. This “double edge” effect combined with the metabolic features of ethanol renders its actions multifaceted. Ethanol is oxidatively metabolized by alcohol dehydrogenase (ADH) or Cyt p450 to acetaldehyde which is next metabolized by aldehyde dehydrogenase (ALDH) to acetate[65]. Phosphatidylethanol (PEth)[66] and fatty acid ethyl esters (FAEE)[67] are generated non-oxidatively from ethanol. Ethanol also causes generation of the reactive oxygen species (ROS) and modulates superoxide dismutases (SOD). Oxidative stress also leads to endoplasmic reticulum (ER) stress resulting in amplification of the injury[68]. It is a combination of these metabolic stresses, including oxidative and non-oxidative, that causes injury to cells (Figure 1) and we term this as Emess.
A function of Emess is dysregulation of methionine metabolism leading to decreased generation of S-adenosylmethionine (SAM)[22,69], a crucial methyl group donating step for DNA and histone methylations[70]. Ethanol feeding affects several enzymes involved in methionine metabolism including a decreased methionine synthetase activity and changes in hepatic SAM, S-adenosyl-L-homocysteine (SAH), SAM/SAH ratio[70,71]. Dysregulation of methionine metabolism is further induced by folate deficiency associated with alcohol abuse[72]. Disturbance in folate metabolism is also related to methylene tetrahydrofolate gene polymorphism[69]. Another part of Emess is glutathione depletion[72]. Glutathione depletion causes both global and regional hypomethylation of DNA[73,74]. SAM administration decreases alcoholic liver injury when given for preventive intervention[22,71]. Although SAM administration improves hepatic function, long term administration of SAM may have deleterious effects because of the accumulation of homocysteine. Betaine supplementation not only maintains SAM levels but also prevents homocysteine accumulation and elevates glutathione levels resulting in amelioration of ethanol-induced hepatic injury[71]. Thus Emess-induced effects on glutathione and methionine levels have profound implications in epigenetic changes.
One of the mechanisms underlying DNA hypomethylation is the direct inhibitory effects of acetaldehyde on enzymes implicated in DNA and histone methylations. Indeed acetaldehyde has been shown to inhibit both DNA methyl transferase[30] and methionine synthase[72,75].
Ethanol metabolism is involved in histone acetylation since inhibitors of alcohol dehydrogenase (4-methyl pyrazole) and aldehyde dehydrogenase (cyanamide) decrease ethanol-induced H3-Lys9 acetylation. This partial effect of inhibitors may imply that part of the ethanol effect on H3 acetylation, may also be independent of its metabolism. Since cyanamide increases the levels of acetaldehyde; and decreases acetylation of histones, acetaldehyde adduct formation is unlikely to account for the observed increases in H3-Lys9 acetylation. Interestingly, treatment of hepatocytes with ethanol metabolite acetate also elicits similar acetylation. Exposure of hepatocytes to acetaldehyde (0.01-1.0 mmol/L) for 24 h also increases H3AcK9. Antioxidant N-acetyl- L-cysteine (NAC, 10 mmol/L) decreases ethanol-induced H3 acetylation by about 50% in rat hepatocytes, suggesting that ROS may play a role in the acetylation[41,43]. Ethanol thus causes characteristic changes in histone acetylation with sensitivity to ethanol metabolic/oxidative stress.
Acute and chronic effects of ethanol on DNA methylation and regional hypermethylation or hypomethylation have yet to be established. Likewise, the effects of ethanol on promoter methylation of repetitive sequences as well as key genes that are implicated in survival and regeneration of liver remain to be explored. A comprehensive investigation into the molecular steps involved in ethanol-induced epigenetic changes and inter-relationships (cross-talks) among epigenetic modifications, i.e. DNA methylations, histone methylations, is warranted. It will be interesting to examine the specificity of the effect of ethanol on individual DNA methyl transferases and histone methyl transferases. The effect of ethanol on histone acetyl transferases[43] or on protein kinases involved in histone phosphorylation[76] has to be ascertained. In parallel, the role of demethylases or deacetylases also needs to be assessed. In therapeutic strategies, drugs which modify the enzymes involved in these pathways can be predicted to alter ethanol-induced tissue damage and should constitute an important goal for future investigations. Additional measures, other than SAM or betaine, to suppress Emess and replenish hepatic glutathione by other agents (e.g. vitamin E, folic acid) should be considered. It must be mentioned here that ethanol-induced epigenetic changes are not limited to liver and GI. Evidence from other systems, e.g. fetal alcohol syndrome[30], neuronal NMDA receptor[77], synuclein[78], brain[79] and HERP gene[80] further emphasizes the importance and potential role of epigenetic changes in alcohol-induced disorders in diverse systems.
Finally, it can be postulated that, as far as ethanol actions are concerned, the ‘epigenetic’ effects of ethanol may be more crucial than its effects on classical ‘genetic alterations’ like DNA deletions or mutations. This remains to be proven. Obviously, epigenetics is set to occupy the center stage of alcoholism research in the next decade.
The authors are thankful to Mr. Daniel Jackson for technical help.
S- Editor Liu Y L- Editor Wang XL E- Editor Bi L
| 1. | Alcohol and Health: Tenth special report to the U.S. Congress on alcohol and health. US DHHS, PHS-NIAAA, 2000. . [Cited in This Article: ] |
| 2. | Room R. Banning smoking in taverns and restaurants--a research opportunity as well as a gain for public health. Addiction. 2005;100:888-890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 3. | Waddington CH. Canalization of development & inheritance of acquired characters. Nature. 1942;150:563-565. [DOI] [Cited in This Article: ] [Cited by in Crossref: 2035] [Cited by in F6Publishing: 2107] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 4. | Cheung P, Lau P. Epigenetic regulation by histone methylation and histone variants. Mol Endocrinol. 2005;19:563-573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 206] [Cited by in F6Publishing: 201] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 5. | De Zhu J. The altered DNA methylation pattern and its implications in liver cancer. Cell Res. 2005;15:272-280. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 6. | Kaneda A, Tsukamoto T, Takamura-Enya T, Watanabe N, Kaminishi M, Sugimura T, Tatematsu M, Ushijima T. Frequent hypomethylation in multiple promoter CpG islands is associated with global hypomethylation, but not with frequent promoter hypermethylation. Cancer Sci. 2004;95:58-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 97] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 7. | Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339-346. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1097] [Cited by in F6Publishing: 1075] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 8. | Lee S, Lee HJ, Kim JH, Lee HS, Jang JJ, Kang GH. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003;163:1371-1378. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 244] [Cited by in F6Publishing: 266] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 9. | Frigola J, Solé X, Paz MF, Moreno V, Esteller M, Capellà G, Peinado MA. Differential DNA hypermethylation and hypomethylation signatures in colorectal cancer. Hum Mol Genet. 2005;14:319-326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 111] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 10. | Kim HC, Roh SA, Ga IH, Kim JS, Yu CS, Kim JC. CpG island methylation as an early event during adenoma progression in carcinogenesis of sporadic colorectal cancer. J Gastroenterol Hepatol. 2005;20:1920-1926. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 47] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Kawakami K, Ruszkiewicz A, Bennett G, Moore J, Grieu F, Watanabe G, Iacopetta B. DNA hypermethylation in the normal colonic mucosa of patients with colorectal cancer. Br J Cancer. 2006;94:593-598. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 113] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 12. | Sato F, Meltzer SJ. CpG island hypermethylation in progression of esophageal and gastric cancer. Cancer. 2006;106:483-493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 116] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 13. | Ogden GR. Alcohol and oral cancer. Alcohol. 2005;35:169-173. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 82] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 14. | La Vecchia C, Lucchini F, Negri E, Levi F. Trends in oral cancer mortality in Europe. Oral Oncol. 2004;40:433-439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 92] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 15. | Ishida E, Nakamura M, Ikuta M, Shimada K, Matsuyoshi S, Kirita T, Konishi N. Promotor hypermethylation of p14ARF is a key alteration for progression of oral squamous cell carcinoma. Oral Oncol. 2005;41:614-622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 53] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 16. | Purohit V, Khalsa J, Serrano J. Mechanisms of alcohol-associated cancers: introduction and summary of the symposium. Alcohol. 2005;35:155-160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 76] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 17. | Mufti SI, Nachiappan V, Eskelson CD. Ethanol-mediated promotion of oesophageal carcinogenesis: association with lipid peroxidation and changes in phospholipid fatty acid profile of the target tissue. Alcohol Alcohol. 1997;32:221-231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 18. | Chang HW, Ling GS, Wei WI, Yuen AP. Smoking and drinking can induce p15 methylation in the upper aerodigestive tract of healthy individuals and patients with head and neck squamous cell carcinoma. Cancer. 2004;101:125-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Nan HM, Song YJ, Yun HY, Park JS, Kim H. Effects of dietary intake and genetic factors on hypermethylation of the hMLH1 gene promoter in gastric cancer. World J Gastroenterol. 2005;11:3834-3841. [PubMed] [Cited in This Article: ] |
| 20. | van Engeland M, Weijenberg MP, Roemen GM, Brink M, de Bruïne AP, Goldbohm RA, van den Brandt PA, Baylin SB, de Goeij AF, Herman JG. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer Res. 2003;63:3133-3137. [PubMed] [Cited in This Article: ] |
| 21. | Yu Z, Kone BC. Hypermethylation of the inducible nitric-oxide synthase gene promoter inhibits its transcription. J Biol Chem. 2004;279:46954-46961. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 22. | Lu SC, Mato JM. Role of methionine adenosyltransferase and S-adenosylmethionine in alcohol-associated liver cancer. Alcohol. 2005;35:227-234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 118] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 23. | Bönsch D, Lenz B, Reulbach U, Kornhuber J, Bleich S. Homocysteine associated genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm. 2004;111:1611-1616. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 106] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 24. | Pöschl G, Stickel F, Wang XD, Seitz HK. Alcohol and cancer: genetic and nutritional aspects. Proc Nutr Soc. 2004;63:65-71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 81] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 25. | Lorincz MC, Groudine M. C(m)C(a/t)GG methylation: a new epigenetic mark in mammalian DNA. Proc Natl Acad Sci USA. 2001;98:10034-10036. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 32] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 26. | Choi MS, Shim YH, Hwa JY, Lee SK, Ro JY, Kim JS, Yu E. Expression of DNA methyltransferases in multistep hepatocarcinogenesis. Hum Pathol. 2003;34:11-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29-39. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 245] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 28. | Ting AH, Jair KW, Schuebel KE, Baylin SB. Differential requirement for DNA methyltransferase 1 in maintaining human cancer cell gene promoter hypermethylation. Cancer Res. 2006;66:729-735. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 58] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 29. | Fang JY, Cheng ZH, Chen YX, Lu R, Yang L, Zhu HY, Lu LG. Expression of Dnmt1, demethylase, MeCP2 and methylation of tumor-related genes in human gastric cancer. World J Gastroenterol. 2004;10:3394-3398. [PubMed] [Cited in This Article: ] |
| 30. | Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcohol Clin Exp Res. 1991;15:395-398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 205] [Cited by in F6Publishing: 214] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 31. | Bönsch D, Lenz B, Fiszer R, Frieling H, Kornhuber J, Bleich S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm. 2006;113:1299-1304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 99] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 32. | Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6156] [Cited by in F6Publishing: 5912] [Article Influence: 246.3] [Reference Citation Analysis (0)] |
| 33. | Dobosy JR, Selker EU. Emerging connections between DNA methylation and histone acetylation. Cell Mol Life Sci. 2001;58:721-727. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 149] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 34. | Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81-120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1456] [Cited by in F6Publishing: 1426] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 35. | de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737-749. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2200] [Cited by in F6Publishing: 2217] [Article Influence: 105.6] [Reference Citation Analysis (0)] |
| 36. | Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263-271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 738] [Cited by in F6Publishing: 696] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 37. | Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905-915. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 615] [Cited by in F6Publishing: 643] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 38. | Li J, Chen P, Sinogeeva N, Gorospe M, Wersto RP, Chrest FJ, Barnes J, Liu Y. Arsenic trioxide promotes histone H3 phosphoacetylation at the chromatin of CASPASE-10 in acute promyelocytic leukemia cells. J Biol Chem. 2002;277:49504-49510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 83] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 39. | Lee DY, Teyssier C, Strahl BD, Stallcup MR. Role of protein methylation in regulation of transcription. Endocr Rev. 2005;26:147-170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 316] [Cited by in F6Publishing: 317] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 40. | Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838-849. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1497] [Cited by in F6Publishing: 1502] [Article Influence: 79.1] [Reference Citation Analysis (0)] |
| 41. | Park PH, Miller R, Shukla SD. Acetylation of histone H3 at lysine 9 by ethanol in rat hepatocytes. Biochem Biophys Res Commun. 2003;306:501-504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 98] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 42. | Lee YJ, Aroor AR, Shukla SD. Temporal activation of p42/44 mitogen-activated protein kinase and c-Jun N-terminal kinase by acetaldehyde in rat hepatocytes and its loss after chronic ethanol exposure. J Pharmacol Exp Ther. 2002;301:908-914. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 44] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 43. | Park PH, Lim RW, Shukla SD. Involvement of histone acetyltransferase (HAT) in ethanol-induced acetylation of histone H3 in hepatocytes: potential mechanism for gene expression. Am J Physiol Gastrointest Liver Physiol. 2005;289:G1124-G1136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 84] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 44. | Kim JS, Shukla SD. Acute in vivo effect of ethanol (binge drinking) on histone H3 modifications in rat tissues. Alcohol Alcohol. 2006;41:126-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 111] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 45. | Kim JS, Shukla SD. Histone h3 modifications in rat hepatic stellate cells by ethanol. Alcohol Alcohol. 2005;40:367-372. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 46. | Mahadev K, Vemuri MC. Ethanol-induced changes in hepatic chromatin and nonhistone nuclear protein composition in the rat. Alcohol. 1998;15:207-211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 47. | Aroor AR, Shukla SD. MAP kinase signaling in diverse effects of ethanol. Life Sci. 2004;74:2339-2364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 153] [Cited by in F6Publishing: 142] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 48. | MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem. 1995;270:11327-11337. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 148] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 49. | Nishigaki M, Aoyagi K, Danjoh I, Fukaya M, Yanagihara K, Sakamoto H, Yoshida T, Sasaki H. Discovery of aberrant expression of R-RAS by cancer-linked DNA hypomethylation in gastric cancer using microarrays. Cancer Res. 2005;65:2115-2124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 129] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 50. | Ray RB, Lagging LM, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438-4443. [PubMed] [Cited in This Article: ] |
| 51. | Calvisi DF, Thorgeirsson SS. Molecular mechanisms of hepatocarcinogenesis in transgenic mouse models of liver cancer. Toxicol Pathol. 2005;33:181-184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 60] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 52. | Nair VD, Yuen T, Olanow CW, Sealfon SC. Early single cell bifurcation of pro- and antiapoptotic states during oxidative stress. J Biol Chem. 2004;279:27494-27501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 77] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 53. | Pruitt K, Ulkü AS, Frantz K, Rojas RJ, Muniz-Medina VM, Rangnekar VM, Der CJ, Shields JM. Ras-mediated loss of the pro-apoptotic response protein Par-4 is mediated by DNA hypermethylation through Raf-independent and Raf-dependent signaling cascades in epithelial cells. J Biol Chem. 2005;280:23363-23370. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 73] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 54. | Edamoto Y, Hara A, Biernat W, Terracciano L, Cathomas G, Riehle HM, Matsuda M, Fujii H, Scoazec JY, Ohgaki H. Alterations of RB1, p53 and Wnt pathways in hepatocellular carcinomas associated with hepatitis C, hepatitis B and alcoholic liver cirrhosis. Int J Cancer. 2003;106:334-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 174] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 55. | Jones MK, Itani RM, Wang H, Tomikawa M, Sarfeh IJ, Szabo S, Tarnawski AS. Activation of VEGF and Ras genes in gastric mucosa during angiogenic response to ethanol injury. Am J Physiol. 1999;276:G1345-G1355. [PubMed] [Cited in This Article: ] |
| 56. | Isayama F, Froh M, Yin M, Conzelmann LO, Milton RJ, McKim SE, Wheeler MD. TNF alpha-induced Ras activation due to ethanol promotes hepatocyte proliferation independently of liver injury in the mouse. Hepatology. 2004;39:721-731. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 57. | Zhang XK, Huang DP, Qiu DK, Chiu JF. The expression of c-myc and c-N-ras in human cirrhotic livers, hepatocellular carcinomas and liver tissue surrounding the tumors. Oncogene. 1990;5:909-914. [PubMed] [Cited in This Article: ] |
| 58. | An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735-748. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 405] [Cited by in F6Publishing: 396] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 59. | Huang S, Litt M, Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 2005;19:1885-1893. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 182] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 60. | Cui R, Nguyen TT, Taube JH, Stratton SA, Feuerman MH, Barton MC. Family members p53 and p73 act together in chromatin modification and direct repression of alpha-fetoprotein transcription. J Biol Chem. 2005;280:39152-39160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 61. | Pogribny IP, James SJ. Reduction of p53 gene expression in human primary hepatocellular carcinoma is associated with promoter region methylation without coding region mutation. Cancer Lett. 2002;176:169-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 59] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 62. | Lin J, Beerm DG. Molecular biology of upper gastrointestinal malignancies. Semin Oncol. 2004;31:476-486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 63. | Yeon JE, Califano S, Xu J, Wands JR, De La Monte SM. Potential role of PTEN phosphatase in ethanol-impaired survival signaling in the liver. Hepatology. 2003;38:703-714. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 90] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 64. | Pani G, Fusco S, Colavitti R, Borrello S, Maggiano N, Cravero AA, Farré SM, Galeotti T, Koch OR. Abrogation of hepatocyte apoptosis and early appearance of liver dysplasia in ethanol-fed p53-deficient mice. Biochem Biophys Res Commun. 2004;325:97-100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 38] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 65. | Dey A, Cederbaum AI. Alcohol and oxidative liver injury. Hepatology. 2006;43:S63-S74. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 398] [Cited by in F6Publishing: 412] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 66. | Shukla SD, Sun GY, Gibson Wood W, Savolainen MJ, Alling C, Hoek JB. Ethanol and lipid metabolic signaling. Alcohol Clin Exp Res. 2001;25:33S-39S. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 67. | Best CA, Laposata M. Fatty acid ethyl esters: toxic non-oxidative metabolites of ethanol and markers of ethanol intake. Front Biosci. 2003;8:e202-e217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 70] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 68. | Ji C, Mehrian-Shai R, Chan C, Hsu YH, Kaplowitz N. Role of CHOP in hepatic apoptosis in the murine model of intragastric ethanol feeding. Alcohol Clin Exp Res. 2005;29:1496-1503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 147] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 69. | Mason JB, Choi SW. Effects of alcohol on folate metabolism: implications for carcinogenesis. Alcohol. 2005;35:235-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 140] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 70. | Halsted CH, Villanueva JA, Devlin AM, Niemelä O, Parkkila S, Garrow TA, Wallock LM, Shigenaga MK, Melnyk S, James SJ. Folate deficiency disturbs hepatic methionine metabolism and promotes liver injury in the ethanol-fed micropig. Proc Natl Acad Sci USA. 2002;99:10072-10077. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 144] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 71. | Kharbanda KK, Rogers DD 2nd, Mailliard ME, Siford GL, Barak AJ, Beckenhauer HC, Sorrell MF, Tuma DJ. A comparison of the effects of betaine and S-adenosylmethionine on ethanol-induced changes in methionine metabolism and steatosis in rat hepatocytes. J Nutr. 2005;135:519-524. [PubMed] [Cited in This Article: ] |
| 72. | Lee TD, Sadda MR, Mendler MH, Bottiglieri T, Kanel G, Mato JM, Lu SC. Abnormal hepatic methionine and glutathione metabolism in patients with alcoholic hepatitis. Alcohol Clin Exp Res. 2004;28:173-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 125] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 73. | Lertratanangkoon K, Wu CJ, Savaraj N, Thomas ML. Alterations of DNA methylation by glutathione depletion. Cancer Lett. 1997;120:149-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 70] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 74. | Fratelli M, Goodwin LO, Ørom UA, Lombardi S, Tonelli R, Mengozzi M, Ghezzi P. Gene expression profiling reveals a signaling role of glutathione in redox regulation. Proc Natl Acad Sci USA. 2005;102:13998-14003. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 138] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 75. | Barak AJ, Beckenhauer HC, Tuma DJ. Methionine synthase. a possible prime site of the ethanolic lesion in liver. Alcohol. 2002;26:65-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 46] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 76. | Lee Y, Shukla SD. Nuclear activation of p38 MAPK and histone H3 phosphorylation by ethanol and acetaldehyde in rat hepatocytes. FASEB J. 2006;25:A1122. [Cited in This Article: ] |
| 77. | Marutha Ravindran CR, Ticku MK. Changes in methylation pattern of NMDA receptor NR2B gene in cortical neurons after chronic ethanol treatment in mice. Brain Res Mol Brain Res. 2004;121:19-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 55] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 78. | Bönsch D, Lenz B, Kornhuber J, Bleich S. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport. 2005;16:167-170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 131] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 79. | Mahadev K, Vemuri MC. Effect of ethanol on chromatin and nonhistone nuclear proteins in rat brain. Neurochem Res. 1998;23:1179-1184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 80. | Bleich S, Lenz B, Ziegenbein M, Beutler S, Frieling H, Kornhuber J, Bönsch D. Epigenetic DNA hypermethylation of the HERP gene promoter induces down-regulation of its mRNA expression in patients with alcohol dependence. Alcohol Clin Exp Res. 2006;30:587-591. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 125] [Article Influence: 6.9] [Reference Citation Analysis (0)] |