Published online Aug 7, 2006. doi: 10.3748/wjg.v12.i29.4640
Revised: October 28, 2005
Accepted: November 10, 2005
Published online: August 7, 2006
AIM: It is known that thyroid hormones alter the bile acid metabolism in humans, however the effect on individual enzymes has been difficult to elucidate. This is mainly due to the lack of human liver cell lines producing bile acids. We used cultures of primary human hepatocytes to study the effects of triiodothyronine (T3) on bile acid synthesis.
METHODS: Primary hepatocytes were isolated from liver tissue obtained from three different patients undergoing liver resection due to underlying malignancy. The hepatocytes were cultured under serum-free conditions and treated from d 1 to d 5 with culture containing 0.1 - 1000 nmol/L of T3. Bile acid formation and mRNA levels of key enzymes were analysed.
RESULTS: The lowest concentration of T3 decreased cholic acid (CA) formation to 43%-53% of controls and chenodeoxycholic acid (CDCA) to 52%-75% of controls on d 5. The highest dose further decreased CA formation to 16%-48% of controls while CDCA formation remained at 50%-117% of controls. Expression of mRNA levels of cholesterol 7α-hydroxylase (CYP7A1) and sterol 12α-hydroxylase (CYP8B1) dose-dependently decreased. Sterol 27-hydroxylase (CYP27A1) levels also decreased, but not to the same extent.
CONCLUSION: T3 dose-dependently decreased total bile acid formation in parallel with decreased expression of CYP7A1 and CYP8B1. CA formation is inhibited to a higher degree than CDCA, resulting in a marked decrease in the CA /CDCA ratio.
- Citation: Ellis ECS. Suppression of bile acid synthesis by thyroid hormone in primary human hepatocytes. World J Gastroenterol 2006; 12(29): 4640-4645
- URL: https://www.wjgnet.com/1007-9327/full/v12/i29/4640.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i29.4640
The effect of thyroid hormone on human bile acid synthesis is still unclear. Several in vivo observations have suggested that thyroid hormone influences the bile acid synthesis[1-3]. However, due to the lack of human in vitro tools such as cell lines synthesizing normal bile acids, investigators have not been able to study this at a cellular level. We have previously shown that primary human hepatocytes cultured under serum-free conditions in the presence of matrixgel (EHS) synthesize and conjugate normal bile acids[4,5]. We also showed that under these conditions they synthesize bile acids with the same capacity and composition as under similar in vivo conditions.
In the conversion of cholesterol into bile acids two main pathways are recognized[6-8]. The neutral pathway is initiated by the 7α-hydroxylation of cholesterol, catalyzed by the microsomal rate-limiting enzyme cholesterol 7α-hydroxylase (CYP7A1). The acidic pathway is initiated by the 27-hydroxylation of cholesterol, catalyzed by the mitochondrial enzyme sterol 27-hydroxylase (CYP27A1). In the acidic pathway the microsomal oxysterol 7α-hydroxylase (CYP7B1) mediates the 7α-hydroxylation. Humans synthesize two primary bile acids, cholic acid (CA) and chenodeoxycholic acid (CDCA). CA has a hydroxyl group at the 12th carbon distinguishing it from CDCA. The hydroxyl group is introduced by the microsomal enzyme sterol 12α-hydroxylase (CYP8B1). Therefore, the total amount of intermediates (from either pathway) that undergoes 12α-hydroxylation determines the ratio of CA to CDCA. Both pathways can synthesize CA and CDCA, however the acidic pathway is believed to produce mainly CDCA. The most important mechanism for regulation of bile acid synthesis is the portal flux of bile acids returning to the liver. The loss of bile acids in the intestine (5%-10%) is compensated by de novo synthesis.
In this study we examined the effect of different doses of triiodothyronine (T3) on bile acid formation and on mRNA levels of CYP7A1, CYP8B1 and CYP27A1 in cultures of primary human hepatocytes.
RPA IIITM kit and pTRI-cyclophilin-Human antisense control template were purchased from Ambion Inc.(Huntingdon, UK). 35S-UTP was purchased from Hartmann Analytic GmbH(Braunschweig, Germany). William’s medium E with glutamax, gentamycin, penicillin-streptomycin, Dulbecco’s phosphate buffered saline, Trizol reagent and Superscript IIRT were purchased from Invitrogen (Täby, Sweden). Actrapid (insulin) 100 IU/mL was from Novo Nordisk Pharma(Malmö, Sweden). Collagenase XI-S (Cat. No. C7557), methylthaizolyldiphenyl-tetrazolium bromide (Cat. No. M5655) and triiodothyronine (Cat. No. T6397) were purchased from Sigma-Aldrich (Sweden). Random hexamers (Cat. No. C1181), recombinant RNasin ribonuclease inhibitor (Cat. No. N2511) and RQ DNase I (Cat. No. M6101) were purchased from Promega (Southampton, UK). Taqman universal master mix (Cat. No. 4304437) and human cyclophilin pre-developed assay (now discontinued and replaced by Human PPIA endogenous control (Cat. No. 4310883E) were purchased from Applied Biosystems (Stockholm, Sweden). Dithiothreitol (Cat. No. 708984) was purchased from Roche(Bromma, Sweden).
Human liver tissue was obtained from three different patients undergoing reduction hepatectomy for metastatic cancer to the liver where normal liver tissue was resected due to the localization of the tumor. Patient A was a 62 year old female, patient B a 47 year old male and patient C a 49 year old female.
Approval to use parts of resected human liver specimens for research was given by the ethics committee at Huddinge University Hospital and the patient’s informed consent was obtained.
Hepatocytes were isolated by a two-step perfusion technique, utilizing EGTA and collagenase as described in detail by Li et al[9]. In brief, liver tissue weighing between 20-150 g was cannulated in 1-2 vessels depending on the size and appearance. The tissue was then perfused with EGTA buffer for about 15 min at a flow rate of 20 mL/min, followed by perfusion with CO2 (5%) buffered solution containing collagenase XI-S (200 mg /L) at 40°Cfor approximately 75 min.
Hepatocytes (3.5 × 106) were cultured under standard conditions in 60 mm culture dishes containing 200 μL EHS matrigel and 3 mL William’s E medium with glutamax, supplemented with insulin (2 IU/L), penicillin G sodium (100 U/mL), streptomycin sulphate (100 μg/mL) and gentamycin (85 μg/mL). Cell culture media were collected and renewed everyday. Three 60 millimeter culture dishes each containing 3.5 × 106 hepatocytes were used for each treatment. The cells and media from three dishes were pooled when harvested. The cells were subjected to different concentrations of T3 (0.1, 1, 10, 100 and 1000 nmol/L) during four days, from d 1 to d 5. On d 5 cell culture media were collected for bile acid analysis and the cells were harvested for RNA isolation and subsequent quantification of specific mRNAs.
Bile acids in cell culture media were analysed as previously described[4]. Briefly, 2 mL of the harvested culture medium, together with deuterium labelled cholic acid (D5) and chenodeoxycholic acid (D4), was subjected to basic hydrolyzation to remove conjugates and the bile acids were extracted using acidic ether. Bile acids were analysed by gas chromatography-mass spectrometry, as described previously[1,10].
Total RNAs were isolated using Trizol reagent as described by the manufacturer. RNA concentration was determined by spectrophotometry at 260 nm. The purity of the RNA was determined by spectrophotometry at 280 nm and the integrity checked by agarose gel electrophoresis stained with ethidium bromide.
mRNA levels of CYP7A1 were determined using quantitative single-plex real time PCR. From each group, 40 μg of total RNA was incubated with 5 U of RQ DNase, 5 μL 10 × RQ buffer in a total volume of 50 μL at 37°C for 20 min, at 20°C for 15 min and finally at 70°C for 10 min. From this, 4 μg of DNase treated RNA was used for cDNA synthesis by mixing together with 2 μL random hexamer primers (100 ng/μL), 1 μL dNTP (10 mmol/L of each) in a total volume of 12 μL. After incubation at 65°C for 5 min, 25°C for 10 min and 42°C for 2 min, 2 μL of DTT (0.1 mol/L), 1 μL of RNasin (40 U/μL), 4 μL 5 × first strand buffer and 1 μL superscript II (200 U/μL) were added. The incubation was continued at 42°C for 50 min and at 70°C for 10 min. For real time PCR 3 μL undiluted cDNA/sample was used in triplicates.
Sequences for CYP7A1 primers and probe were kindly provided by Dr. Hermansson, AstraZeneca, Mölndal, Sweden. Reversed primer is: 5’AGA GCA CAG CCC AGG TAT G 3’; forward primer, 5’ TGA TTT GGG GGA TTG CTA TA 3’ (300 mmol/L); and probe 5’ TGG TTC ACC CGT TTG CCT TCT CCT 3’ (200 mmol/L). The taqman probe was labelled with FAM and TAMRA. The assays were performed according to ABI user bulletin #2 protocol in a total volume of 25 μL. VIC labelled human cyclophilin pre-developed taqman assay from Applied Biosystems was used for internal control. The assay was performed and analysed using ABI Prism 7700 sequence detector system.
Abundance of specific mRNA was quantified using RPA IIITM. cRNA probes were synthesized using T3- or T7-polymerase and labelled with 35S-UTP. Cyclophilin was used as an internal standard and gave a protected fragment size of 103 bp.
The protected fragment size of the probe for CYP27A1 was 297 bp and for CYP8B1 442 bp. The assay was performed according to the manufacturer’s protocol. In brief, 10-15 μg of total RNA was co-precipitated in 2.5 volumes of ethanol and 0.5 mol/L NH4OAc with about 80 000 cpm of each probe at -20°C for 15 min. Following centrifugation at 15 000 rpm for 15 min the pellets were dissolved in 10 μL hybridisation buffer, heated to 95°C for 3 min and incubated at 46°C overnight. The hybridised RNA samples were then treated with RNase A and RNase A/T1 diluted 1:50 in RNase solution at 37°C and inactivated after 30 min by adding inactivation/precipitation solution. Following precipitation at -20°C for 15 min the samples were centrifuged at 15 000 r/min for 15 min and the pellets were air dried and dissolved in 10 μL of loading buffer.
The protected fragments were separated on a denaturing 5% polyacrylamide urea gel and detected and quantified using a Fuji BAS 1800 Phospho-imager.
The significance of differences between groups for the mRNA analysis was tested by a one-way ANOVA followed by a post-hoc test. To stabilize the variances, data were logarithmically transformed. Due to variances between patients, statistical analysis of the bile acid formation could not be performed.
We have analyzed the purity of the preparation of hepatocytes that we plated. Following the final low speed centrifugation step the cell pellet was resuspended in plating media and samples of the cell preparation were spotted onto microscopic glass slides, air dried and fixed in formalin. Preparations from 2 separate human cases were analyzed. Duplicate slides were exposed to antibodies for low molecular weight cytokeratins (AE1/AE3), albumin or hepatocyte specific antibody HEPR. The results showed that 93% of the cells in the preparation reacted with antibodies to cytokeratin, albumin and HEPR, indicating that 93% of the cells plated on culture dishes were epithelial (cytokeratin positive) and parenchymal hepatocytes (HEPR and albumin positive). Most of the remaining cells were small and appeared to be hematopoietic. Hematopoietic cells would not be expected to attach to culture dishes and would be removed by media changes.
The cells were treated for 4 d (from d 1 to d 5) with different concentrations of T3. The cell culture media were changed daily and collected on d 5 of culture after 24 h incubation. In control cells the total bile acid formation was 1515 ng/mL of culture media (920 ng CA and 595 ng CDCA per mL) and the CA/CDCA ratio 1.5. Treatment with 0.1 nmol/L of T3 decreased total formation by 53% to 710 ng/mL and lowered the CA/CDCA ratio to 1.3 (Table 1). Higher doses of T3 decreased bile acid formation even further in a dose dependent manner, down to 36% of control with a CA/CDCA ratio of 0.8 for the highest dose, 1000 nmol/L.
| Patient | Addition of T3 | CA | CDCA | Ratio |
| ng/mL | Medium/24 h | CA/CDCA | ||
| A | Control | 920 | 595 | 1.5 |
| 0.1 nmol/L | 400 | 310 | 1.3 | |
| 1 nmol/L | 375 | 375 | 1.0 | |
| 10 nmol/L | 340 | 390 | 0.9 | |
| 100 nmol/L | 250 | 285 | 0.9 | |
| 1 μmol/L | 245 | 295 | 0.8 | |
| B | Control | 280 | 60 | 4.7 |
| 0.1 nmol/L | 135 | 45 | 3.0 | |
| 1 nmol/L | 95 | 70 | 1.4 | |
| 10 nmol/L | 55 | 60 | 0.9 | |
| 100 nmol/L | 45 | 65 | 0.7 | |
| 1 μmol/L | 45 | 70 | 0.6 | |
| C | Control | 805 | 502 | 1.6 |
| 0.1 nmol/L | 430 | 345 | 1.2 | |
| 1 nmol/L | 477 | 392 | 1.2 | |
| 10 nmol/L | 395 | 425 | 0.9 | |
| 100 nmol/L | 372 | 405 | 0.9 | |
| 1 μmol/L | 382 | 420 | 0.9 |
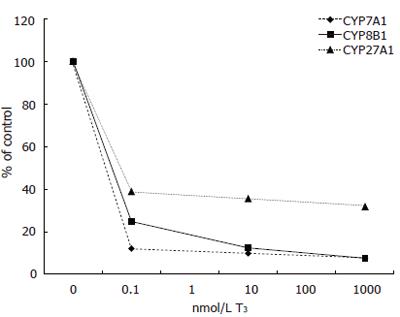
Expression of CYP7A1 mRNA levels was decreased by all doses of T3 tested. Addition of 0.1 nmol/L of T3 decreased CYP7A1 mRNA levels from 4.45 to 0.54 (arbituary value) (12% of control), 10 nmol/L to 0.44 and 1000 nmol/L further down to 0.34 (8% of control). Two of the measure points were lost due to human error (Figure 1).
Similarly, expression of CYP8B1 mRNA decreased with T3 treatment. Addition of 0.1 nmol/L of T3 decreased CYP8B1 from 0.226 to 0.056 (arbituary value) (12% of control), 10 nmol/L to 0.028 and 1000 nmol/L to 0.017 (8% of control) (Figure 1).
Also CYP27A1 mRNA levels decreased with T3 treatment. Addition of 0.1 nmol/L lowered CYP27A1 levels from 1.050 to 0.408 (arbituary value) (39% of control), 10 nmol/L to 0.373 and 1000 nmol/L to 0.334 (32% of control) (Figure 1).
The bile acid formation in hepatocytes from patient B was much lower than in cells from patients A and C. In control cells the total bile acid formation was 340 ng/mL of culture media (280 ng CA and 60 ng CDCA per mL) and the CA/CDCA ratio was 4.7. Treatment with 0.1 nmol/L of T3 decreased total formation by 47% to 180 ng/mL, and lowered CA/CDCA ratio to 3.0 (Table 1). Higher doses of T3 decreased bile acid formation even further down to 34% of control and CA/CDCA ratio to 0.6 for the highest dose, 1000 nmol/L.
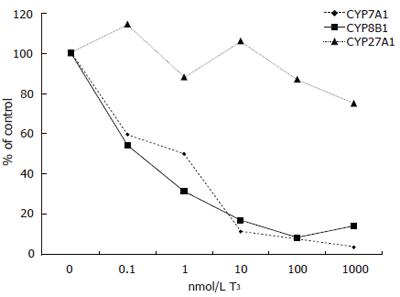
Expression of CYP7A1 mRNA levels was decreased by all doses of T3 tested but not to the same extent as in cells from patient A. Addition of 0.1 nmol/L of T3 decreased CYP7A1 mRNA levels from 0.745 to 0.444 (arbituary value) (60% of control), 1 nmol/L to 0.373, 10 nmol/L to 0.086, 100 nmol/L to 0.063 and 1000 nmol/L further down to 0.028 (4% of control) (Figure 2).
Similarly, expression of CYP8B1 decreased with T3 treatment. Addition of 0.1 nmol/L of T3 decreased CYP8B1 mRNA levels from 0.175 to 0.095 (arbituary value) (54% of control), 1 nmol/L to 0.055, 10 nmol/L to 0.030, 100 nmol/L to 0.015 and 1000 nmol/L further down to 0.025 (14% of control) (Figure 2).
T3 had a modest effect on the expression of CYP27A1 in patient B. Addition of 0.1 nmol/L of T3 changed CYP27A1 mRNA levels from 0.420 to 0.480 (arbituary value) (114% of control), 1 nmol/L to 0.370, 10 nmol/L to 0.445, 100 nmol/L to 0.365 and 1000 nmol/L down to 0.315 (75% of control) (Figure 2).
In control cells total bile acid formation was 1308 ng/mL of culture media (805 ng CA and 502 ng CDCA per mL) and the CA/CDCA ratio was 1.6. Treatment with 0.1 nmol/L of T3 decreased the total formation by 41% to 775 ng/mL and lowered the CA/CDCA ratio to 1.2 (Table 1). Higher doses of T3 decreased bile acid formation to 61% of control and lowered the CA/CDCA further down to 0.9 for the highest dose, 1000 nmol/L.
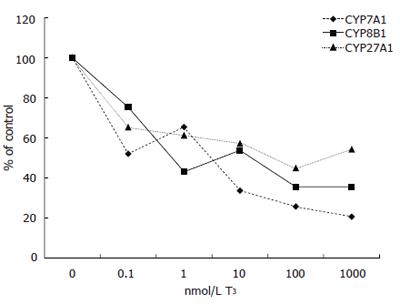
Expression of CYP7A1 mRNA levels was decreased by all doses of T3 tested, but not as strongly as in patients A and B. Addition of 0.1 nmol/L of T3 decreased CYP7A1 mRNA levels from 3.15 to 1.64 (arbituary value) (52% of control), 1 nmol/L to 2.06, 10 nmol/L to 1.06, 100 nmol/L to 0.81 and 1000 nmol/L further down to 0.65 (21% of control) (Figure 3).
Similarly, expression of CYP8B1 mRNA decreased with T3 treatment. Addition of 0.1 nmol/L of T3 decreased CYP8B1 mRNA levels from 0.093 to 0.070 (arbituary value) (75% of control), 1 nmol/L to 0.040, 10 nmol/L to 0.050, 100 nmol/L and 1000 nmol/L both to 0.033 (35% of control) (Figure 3).
Also CYP27A1 mRNA levels decreased with T3 treatment. Addition of 0.1 nmol/L lowered CYP27A1 levels from 0.245 to 0.160 (arbituary value) (65% of control), 1 nmol/L to 0.150, 10 nmol/L to 0.140, 100 nmol/L to 0.110 and 1000 nmol/L to 0.133 (54% of control) (Figure 3).
Following logarithmical transformation, statistical analysis of mRNA levels from all three patients showed that the highest dose (1000 nmol/L) of T3 significantly decreased CYP7A1 mRNA levels (P < 0.05). CYP8B1 mRNA levels was significantly decreased by 10 and 100 nmol/L of T3. Treatment with T3 did not significantly decrease CYP27A1 mRNA levels.
In accordance with our previous studies[4,5,11,12], CA and CDCA, constituted more than 95% of total sterols and the ratio between the two bile acids averaged 2.6.
T3 dose-dependently decreased formation of bile acids to 44% of controls. The reduction of CA formation was greater than the decline in CDCA formation and resulted in a marked decrease in the CA/CDCA ratio. These results are in reasonable agreement with previous in vivo studies. Angelin et al[13] studied bile acid metabolism in hypothyroid patients before and during replacement therapy to euthyroid state and found that CDCA synthesis was stimulated by approximately 40%, whereas CA did not change with hormone treatment. However, the relative concentration of deoxycholic acid (DCA) in bile decreased from 30% to 19% and CDCA concomitantly increased with hormone therapy. This was confirmed by Kosuge et al[2] who concluded that the characteristic effect of thyroid hormone on serum bile acid composition in man is the shift from the “family” of cholic to that of CDCA by showing that DCA is the most prominent bile acid in hypothyroid patients and that CDCA is the most prominent bile acid in hyperthyroid patients. Paulezki et al[3] studied bile acid metabolism in hyperthyroid patients before and during treatment to euthyroid state and showed that thyroid hormone excess caused a 34% reduction in CA synthesis with no apparent change of CDCA synthesis and a 20% decrease in total bile acid synthesis. The authors suggested that thyroid hormone inhibited hepatic 12α-hydroxylation.
In contrast to this, Sauter et al failed to show any effect of T3 treatment on the serum levels of the bile acid intermediate 7α-hydroxy-4-cholestene-3-one (C4) (believed to reflect CYP7A1 activity in vivo) in hypo- and hyperthyroidism before and after treatment to euthyroid state[14]. A recent study by Galman et al showed that there are marked diurnal changes of C4 levels during the day[15]. The authors could also see a weak but significant correlation of C4 with cholesterol and therefore corrected C4 levels for cholesterol. These findings make it difficult to evaluate the results of the C4 levels of the mentioned study. Also, the fact that C4 is the main substrate for CYP8B1 and that one study suggested that CYP8B1 might be inhibited by thyroid hormone needs to be considered.
The mRNA levels of the rate-limiting enzyme CYP7A1 in the neutral pathway were also dose-dependently decreased by T3. Our data support the human promoter studies by Wang et al who found that thyroid hormone repressed CYP7A1 activity[16]. Furthermore, Drover et al found a distinct thyroid hormone response element that mediates T3 repression in the human CYP7A1 promoter[17]. This is in contrast to the study in rats where Crestani et al failed to demonstrate an effect of thyroid hormone on the Cyp7a1 promoter activity[18]. However addition of 1 μmol/L of T4 to primary rat hepatocytes increased the transcriptional activity of Cyp7a1 by 3.7 fold but did not alter steady state mRNA levels[19].
In the present study we did not measure the enzyme activity of CYP7A1 because of lack of material, but since the effects on the mRNA levels were in agreement with the effects on the bile acid formation we believed that it would be unlikely that T3 would have an opposite effect on the enzyme activity.
Similar to CYP7A1, the CYP8B1 mRNA levels were also dose-dependently decreased by T3. Although previous in vivo data pointed towards an effect on CYP8B1, this is to our knowledge the first report of the effects of T3 on human CYP8B1 mRNA levels. In rats however, T3 has been shown to suppress both Cyp8b1 activity and mRNA levels[20].
Treatment with T3 also decreased CYP27A1 levels, but to a much lower degree than CYP7A1 and CYP8B1. This finding is in line with the contention that CYP27A1 is regulated to a lower degree than CYP7A1 in humans, as has been shown in connection with bile acid mediated feedback regulation[21]. According to a previous study by Stravitz et al[22] Cyp27a1 is not regulated by thyroid hormone at all in rat.
In a previous study[4] we investigated the effect of the combination of T3 and dexamethasone on bile acid synthesis in rat vs human hepatocytes. Since it was previously shown that T3 and dexamethasone had a synergistic effect on Cyp7a1 mRNA levels in primary rat hepatocytes[19] and since neither T3 nor dexamethasone alone had any effect in rat hepatocytes, we did not focus on the effect of T3 or dexamethasone alone in primary human hepatocytes. However, we could see that T3 and dexamethasone alone tended to decrease bile acid formation but had no consistent effect on CYP7A1 mRNA levels (data not shown). The results of the present study and the fact that combination of T3 and dexamethasone did not change human bile acid formation or CYP7A1 mRNA levels in our prior study, suggest that dexamethasone might have an effect on human bile acid synthesis. This is very interesting and needs to be elucidated in future studies.
The concentrations of T3 used in this study ranged from 0.1 nmol/L to 1000 nmol/L. The highest dose was included because this is a dose commonly used in combination with dexamethasone in cultures of primary rat hepatocytes. Reduction of methylthaizolyldiphenyl-terazolium bromide (MTT) showed no signs of cytotoxicity in the dose range used here (data not shown). It should be emphasized that the most dramatic effects were seen with the lowest T3 concentration, supporting that the effects observed are physiological.
In conclusion, it has been shown that T3 dose-dependently decreases bile acid formation and expression levels of CYP7A1 and CYP8B1 mRNAs in primary human hepatocytes. CYP27A1 is also decreased, but to a lower degree. CA formation is inhibited to a higher degree than CDCA formation, resulting in a marked decrease in the CA/CDCA ratio.
The author would like to thank Professor Curt Einarsson and Professor Ingemar Björkhem for critical discussions regarding the manuscript. The skilful technical assistance of Mrs. Anita Lövgren-Sandblom regarding the GC/MS analysis is gratefully acknowledged.
S- Editor Wang J L- Editor Zhu LH E- Editor Bai SH
| 1. | Angelin B, Bjökhem I, Einarsson K. Individual serum bile acid concentrations in normo- and hyperlipoproteinemia as determined by mass fragmentography: relation to bile acid pool size. J Lipid Res. 1978;19:527-537. [PubMed] [Cited in This Article: ] |
| 2. | Kosuge T, Beppu T, Kodama T, Hidai K, Idezuki Y. Serum bile acid profile in thyroid dysfunction and effect of medical treatment. Clin Sci (Lond). 1987;73:425-429. [PubMed] [Cited in This Article: ] |
| 3. | Pauletzki J, Stellaard F, Paumgartner G. Bile acid metabolism in human hyperthyroidism. Hepatology. 1989;9:852-855. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 22] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 4. | Ellis E, Goodwin B, Abrahamsson A, Liddle C, Mode A, Rudling M, Bjorkhem I, Einarsson C. Bile acid synthesis in primary cultures of rat and human hepatocytes. Hepatology. 1998;27:615-620. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 42] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Axelson M, Ellis E, Mörk B, Garmark K, Abrahamsson A, Björkhem I, Ericzon BG, Einarsson C. Bile acid synthesis in cultured human hepatocytes: support for an alternative biosynthetic pathway to cholic acid. Hepatology. 2000;31:1305-1312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 50] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | Russell DW. Cholesterol biosynthesis and metabolism. Cardiovasc Drugs Ther. 1992;6:103-110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 137] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 7. | Princen HM, Post SM, Twisk J. Regulation of Bile Acid Biosynthesis. Current Pharmaceutical Design. 1997;3:59-84. [Cited in This Article: ] |
| 8. | Vlahcevic ZR, Pandak WM, Stravitz RT. Regulation of bile acid biosynthesis. Gastroenterol Clin North Am. 1999;28:1-25, v. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 121] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 9. | Li AP. Roque MA, Beck DJ, Kaminski DL. Isolation and culturing of hepatocytes from human livers. J Tiss Cult Meth. 1992;14:139-146. [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 77] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 10. | Björkhem I, Falk O. Assay of the major bile acids in serum by isotope dilution-mass spectrometry. Scand J Clin Lab Invest. 1983;43:163-170. [PubMed] [Cited in This Article: ] |
| 11. | Einarsson C, Ellis E, Abrahamsson A, Ericzon BG, Bjorkhem I, Axelson M. Bile acid formation in primary human hepatocytes. World J Gastroenterol. 2000;6:522-525. [PubMed] [Cited in This Article: ] |
| 12. | Ellis E, Axelson M, Abrahamsson A, Eggertsen G, Thörne A, Nowak G, Ericzon BG, Björkhem I, Einarsson C. Feedback regulation of bile acid synthesis in primary human hepatocytes: evidence that CDCA is the strongest inhibitor. Hepatology. 2003;38:930-938. [PubMed] [Cited in This Article: ] |
| 13. | Angelin B, Einarsson K, Leijd B. Bile acid metabolism in hypothyroid subjects: response to substitution therapy. Eur J Clin Invest. 1983;13:99-106. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 23] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 14. | Sauter G, Weiss M, Hoermann R. Cholesterol 7 alpha-hydroxylase activity in hypothyroidism and hyperthyroidism in humans. Horm Metab Res. 1997;29:176-179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 15. | Galman C. Modulation of bile acid and cholesterol metabolism in health and diease [Doctoral]. Stockholm: Karolinska Institute 2004; 1-35. [Cited in This Article: ] |
| 16. | Wang DP, Stroup D, Marrapodi M, Crestani M, Galli G, Chiang JY. Transcriptional regulation of the human cholesterol 7 alpha-hydroxylase gene (CYP7A) in HepG2 cells. J Lipid Res. 1996;37:1831-1841. [PubMed] [Cited in This Article: ] |
| 17. | Drover VA, Wong NC, Agellon LB. A distinct thyroid hormone response element mediates repression of the human cholesterol 7alpha-hydroxylase (CYP7A1) gene promoter. Mol Endocrinol. 2002;16:14-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 18. | Crestani M, Stroup D, Chiang JY. Hormonal regulation of the cholesterol 7 alpha-hydroxylase gene (CYP7). J Lipid Res. 1995;36:2419-2432. [PubMed] [Cited in This Article: ] |
| 19. | Hylemon PB, Gurley EC, Stravitz RT, Litz JS, Pandak WM, Chiang JY, Vlahcevic ZR. Hormonal regulation of cholesterol 7 alpha-hydroxylase mRNA levels and transcriptional activity in primary rat hepatocyte cultures. J Biol Chem. 1992;267:16866-16871. [PubMed] [Cited in This Article: ] |
| 20. | Andersson U, Yang YZ, Björkhem I, Einarsson C, Eggertsen G, Gåfvels M. Thyroid hormone suppresses hepatic sterol 12alpha-hydroxylase (CYP8B1) activity and messenger ribonucleic acid in rat liver: failure to define known thyroid hormone response elements in the gene. Biochim Biophys Acta. 1999;1438:167-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 21. | Björkhem I, Araya Z, Rudling M, Angelin B, Einarsson C, Wikvall K. Differences in the regulation of the classical and the alternative pathway for bile acid synthesis in human liver. No coordinate regulation of CYP7A1 and CYP27A1. J Biol Chem. 2002;277:26804-26807. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 22. | Stravitz RT, Vlahcevic ZR, Russell TL, Heizer ML, Avadhani NG, Hylemon PB. Regulation of sterol 27-hydroxylase and an alternative pathway of bile acid biosynthesis in primary cultures of rat hepatocytes. J Steroid Biochem Mol Biol. 1996;57:337-347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 65] [Article Influence: 2.3] [Reference Citation Analysis (0)] |