Published online Feb 15, 2004. doi: 10.3748/wjg.v10.i4.598
Revised: September 18, 2003
Accepted: October 23, 2003
Published online: February 15, 2004
AIM: Only a minority of patients carrying a defined viral aetiologic agent develop cirrhosis and ultimately hepatocellular carcinoma (HCC), the mechanism underlying the worsening is still undefined. Experimental infection by Helicobacter hepaticus in mice causes chronic hepatitis and HCC and recently, more Helicobacter species (Helicobacter spp.) have been detected in the liver of patients suffering from cholestatic diseases and HCC arising from non-cirrhotic liver. We investigated whether Helicobacter spp. sequences could be detected in the liver of patients with cirrhosis and HCC compared to subjects with metastasis to liver from colon cancer.
METHODS: Twenty-three liver samples from patients operated upon for HCC superimposed on hepatitis C virus (HCV)-related cirrhosis and 6 from patients with resected metastases from colorectal cancer, were tested by polymerase chain reaction for presence of genomic 16S rRNA of Helicobacter genus using specific primers. DNA sequencing and cag A gene analysis were also performed.
RESULTS: Genomic sequences of Helicobacter spp. were found in 17 of 20 (85%) liver samples from patients with HCC and in 2 of 6 samples from patients with liver metastasis. In three samples of the first group the result was uncertain. H pylori was revealed in 16 out of 17 positive samples and Helicobacter pullorum in the other.
CONCLUSION: Helicobacter spp., carcinogenic in mice, were found at a higher frequency in the liver of patients with HCV-related cirrhosis and HCC than those in patients without primary liver disease.
- Citation: Pellicano R, Mazzaferro V, Grigioni WF, Cutufia MA, Fagoonee S, Silengo L, Rizzetto M, Ponzetto A. Helicobacter species sequences in liver samples from patients with and without hepatocellular carcinoma. World J Gastroenterol 2004; 10(4): 598-601
- URL: https://www.wjgnet.com/1007-9327/full/v10/i4/598.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i4.598
Hepatocellular carcinoma (HCC) is the fourth cause of cancer death worldwide[1]. This tumor often follows chronic liver inflammation and longstanding cirrhosis. Cirrhosis of the liver is a diffuse process, characterised by fibrosis and nodule formation, which stems from hepatocellular injury. The cellular necrosis might originate from viral, toxic, metabolic and autoimmune sources[2]. Moreover, hepatitis B and hepatitis C viruses (HBV and HCV) are well-known etiologic factors for liver cirrhosis, and have been classified by the International Agency for Research on Cancer (IARC) as type I liver carcinogens. Both viruses are highly prevalent in the adult population in Italy[3,4]. Both viral and host characteristics have been reported to influence the outcome of HBV or HCV infection and the development of cirrhosis, but they do not explain all the epidemiological variations of the disease. For instance, in the case of HCV, the viral genotype 1b[5] has been repeatedly connected with severity of liver outcome, although in other reports this issue has been debated[6]. Host characteristics also play an important role in the pathogenesis of liver disease. For example, HLA class II haplotype DR5 has been shown to protect patients from the development of cirrhosis[7].
Chronic inflammation per se is known to be a factor of progression towards cancer[8], though HCV in itself does not appear to foster a strong inflammatory mechanism[9]. It is thus not yet clear what factors most influence the outcome of the liver disease. Searching for other noxae has therefore become mandatory. A new infectious agent, Helicobacter hepaticus (H. hepaticus), that causes chronic active hepatitis and associated liver tumors in A/J Cr mice, has been described by Ward et al[10]. Recently, other Helicobacter species (Helicobacter spp.), including Helicobacter pylori (H pylori), bacteria associated with the pathogenesis of gastric[11-14] and extradigestive manifestations[15,16], have been detected in the liver of persons suffering from cholestatic diseases and HCC arising from non-cirrhotic liver[17,18]. We have previously reported a high frequency of H pylori and Helicobacter pullorum (H.pullorum) sequences detected by the polymerase chain reaction (PCR), in the liver of patients with cirrhosis and superimposed HCC[19].
To demonstrate a role of the bacteria as a co-factor in the development of end-stage of liver disease in humans, it is fundamental to evidence whether Helicobacter occurs at the same frequency in the liver of patients without primary liver disease. The present study investigated whether Helicobacter spp. genomes could be detected in liver specimens from patients without any diagnosis of chronic active hepatitis, cirrhosis or HCC, compared with specimens from HCC patients. Therefore, we analysed liver samples from subjects operated upon for metastasis to the liver from colon cancer and, the presence of Helicobacter was assayed by molecular methods.
Liver samples were obtained during the surgical excision for cure from 23 patients operated upon for HCC superimposed on hepatitis C virus (HCV)-related cirrhosis and from 6 patients suffering from metastatic cancer to the liver, in whom surgery might obtain remission of the disease. All samples were immediately frozen in liquid nitrogen and stored at -80 °C prior to testing. The architectural pattern and histologic grade of tumor differentiation of HCC were recorded in accordance with World Health Organisation (WHO) proposals[20]. HCV infection was demonstrated by means of an ELISA (Axsym System, Abbott Diagnostics) and an RIBA (RIBA II, LIA, HCV3, Sorin, Saluggia, Italy), and confirmed through detection of circulating HCV-RNA in a PCR.
Approximately 3 mm3 of tissue was cut and incubated with 0.4 mg/ml proteinase K (Sigma, St Louis) in 0.5 ml extraction buffer (10 mM Tris-HCl, pH 7.5; 1 mM ethylene diamine tetra acetate or EDTA, pH 8; 0.25 M sodium chloride; 0.2% sodium dodecyl sulphate or SDS) at 56 °C on an agitator until total tissue digestion, the proteinase K was inactivated by boiling it for 10 min. The mixture was centrifuged at 14000 rpm (Eppendorf centrifuge 5415 C) at room temperature, and to the supernatant, 1 volume of phenol/chloroform (1:1 ratio, equilibrated with 0.1 M phosphate buffered saline, pH 7.5) was added. After vortexed and centrifuged at 14000 rpm, the aqueous phase was precipitated with 2 volumes of 100% ethanol containing 10% volume of 3 M sodium acetate, pH 4.8 at -80 °C for 20 min or at -20 °C overnight. After two washes with 75% ethanol, the pellet was dried and resuspended in 300 μl TE buffer (10 mM Tris-HCl, pH 7.5, 5 mM EDTA, pH 8) for further use.
Amplification of 16S rRNA gene Samples were amplified by Helicobacter genus-specific 16S ribosomal RNA(16S rRNA) primers (designated Heli, Table 1) that could identify genomic sequences of 26 species of Helicobacter.
| Primers | Sequences 5’→3’ |
| Heli-nest.S | 5’ ATT AGT GGC GCA CGG GTG AGT AA 3’ |
| Heli-nest-R | 5’ TTT AGC ATC CCG ACT TAA GGC 3’ |
| Heli-S | 5’ GAA CCT TAC CTA GGC TTG ACA TTG 3’ |
| Heli-R | 5’ GGT GAG TAC AAG ACC CGG GAA 3’ |
| CagA-1 | 5’ GGT GAG TAC AAG ACC CGG GAA 3’ |
| CagA-2 | 5’TTA GAA TAA TCA ACA AAC ATC ACG CCA T 3’ |
First amplification step Genomic DNA was amplified in a total volume of 50 μl containing PCR buffer (1X), 200 µM dNTP, 4 mM Mg 2+, 0.2 μM primers (Heli-nest-S and Heli-nest-R, Table 1), 1 U Taq DNA polymerase (GibcoBRL, USA) and 1 μg of total DNA. The reaction mixture was initially denatured for 2 min at 94 °C then amplified for 35 cycles as follows: denaturation for 30 s at 94 °C, primer annealing for 30 s at 55 °C and extension for 1.5 min at 72 °C. A final extension step was done for 5 min at 72 °C (Thermocycler HYBAID–OmniGene TR35M2-UK). An amplification product of 1300 bp by the forward and reverse primers was expected.
Second amplification step PCR was repeated as above with minor alterations: 1 μl of amplicon was used from the first amplification step, the annealing temperature was brought to 60 °C, extension time lasted 30 s for 35 cycles and Heli-R/ Heli-S primers (Table 1) were used. The expected size of the amplicon was 480 bp.
Samples generating a positive result from the Nested-PCR were subsequently analysed with a different pair of primers (CagA, Table 1). The CagA-1 and CagA-2 primers used (Table 1) amplified a 290 bp product from the 128 KDa Cag A protein, typical of H pylori[21]. Fifty microliter of reaction mixture was prepared containing PCR buffer (1X), 2 mM of Mg 2+, 200 μM dNTP, 0.2 μM primers, 1 U of Taq DNA polymerase and 1 μg of DNA. Amplification consisted of an initial denaturation for 2 min at 94 °C then amplified for 50 cycles as follows: for 30 s at 94 °C, for 30 s at 61 °C and for 1 min at 72 °C. Extension was continued for another cycle for 5 min at 72 °C. The PCR products were analysed on a 2% agarose gel. Positive and negative controls were included. VCS Hae III and VCS Hinf I were used as DNA size standard for amplification with Heli primers and Cag A primers, respectively.
For this purpose, 10 μl of the amplified DNA samples was electrophoresed on a 1% agarose gel. The samples were then transferred to nylon hybond N+ membrane (Amersham Pharmacia Biotech Inc., UK) treated with denaturing agent (0.5 M sodium hydroxide, 1.5 M sodium chloride). The membrane was then neutralised with 5X SSC (0.75 M sodium chloride and 0.075 M sodium acetate) for 1 min. The membrane was prehybridised in buffer (5X SSPE (0.6 M NaCl, 40 mM NaH2PO4, 4 mM EDTA), 0.5% SDS, 5X Denhardt’s solution, 0.02 mg/ml Salmon sperm) at 65 °C for 4 h. A freshly denatured probe, amplified from H. pullorum DNA and labelled with P32(RediprimeTM II, random labelling system, Amersham Pharmacia Biotech Inc., UK) according to the manufacturer’s instructions, was purified using MicrospinTM S-400 (HR Columns, Amersham Pharmacia Biotech Inc., New Jersey, USA). Hybridisation was performed overnight at 65 °C in hybridisation solution containing blocking reagent (5X SSPE, 0.5% SDS, 5X Denhardt’s solution) and the DNA probe. The nylon membrane was then washed in 2X SSC, 0.1% SDS for 15 min at RT, 2X SSC, 0.1% SDS for 15 min at 65 °C, 1X SSC, 0.1% SDS for 15 min at 65 °C and 0.5X SSC, 0.1% SDS for 10 min at 65 °C. The bound probes were finally detected by autoradiography after 4 h at RT (Kodak scientific imaging film, X-OMAT/AR, Rochester, New York).
PCR-amplified Helicobacter genus-specific PCR products were run on a 2% agarose gel, purified using the QIAquick gel extraction kit (QIAGEN, Germany) and sequence analysis was performed with an Applied Biosystems DNA sequencer (Perkin-Elmer, Applied Biosystems, Warrington, Great Britain) following the procedure of the manufacturer, using the ABI PRISM 310 big dye terminator cycle sequencing kit. The sequences were entered, aligned and compared with known Helicobacter species using the BLAST programme (Genetics Computer Group, Madison, Wis.).
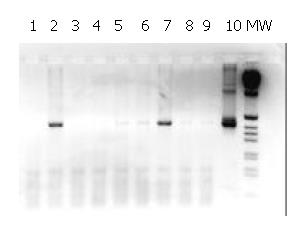
We found DNA sequences typical of Helicobacter spp. in 17 of 20 (85%) livers from patients operated for HCC using Helicobacter genus-specific 16S rRNA primers. Three samples (15%) were negative, while in the remaining 3 out of 23, the results were uncertain. Two of 6 liver samples obtained from 6 patients whose livers were resected for metastasis due to colon cancer, contained sequences typical of Helicobacter spp.
The 480 bp product obtained in the second round of the nested-PCR with Heli primers is shown in Figure 1. The liver samples with a positive result by this method were all positive by Southern blot hybridisation using P32-labelled probe generated from H.pullorum DNA (data not shown). These results confirmed the presence of Helicobacter gene sequences in the liver samples analysed from patients with cirrhosis and HCC.
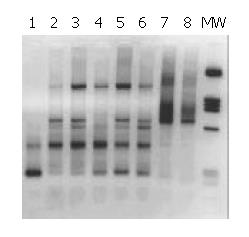
The patients whose liver samples gave a signal typical for Helicobacter genus with Heli primers (Table 1), were analysed with primers specific for the amplification of a 290 bp stretch of the cag A gene[21]. The bands amplified corresponded to the expected size (Figure 2).
Overall, 16 out of 17 (94%) liver tissues from patients who were positive when tested for Helicobacter genus, were also found to be positive when tested for the presence of a sequence that was typical only for type I Helicobacter pylori, i.e., the cag A gene.
The Helicobacter positive DNA fragments were sequenced. Comparison of DNA sequences of the genomic 16S rRNA amplicons with those of known Helicobacter spp. was performed using sequence alignment with the BLAST programme. The presence of H pylori and H. pullorum sequences was thus confirmed in these liver samples.
Despite the profound impact of HCC on human health worldwide[22], its pathogenesis remains uncertain. Thanks to the availability of very efficient vaccines that could prevent HBV infection, WHO has sought to eradicate HBV infection and hence its long-term consequences such as HCC. However, in most industrialised countries, it has been shown that neither HBV nor Schistosoma infections play a major role in the development of HCC, instead, the majority of patients carry HCV. This virus does not integrate into host DNA, and it is likely that the mechanism of carcinogenesis differs from that of HBV. Therefore, the need to clarify the pathogenic mechanisms by which HCV may lead to liver cancer has become of prime importance.
Since the currently known risk factors cannot explain all aspects of its progression to HCC, other causal mechanisms should be explored[23]. From an epidemiological aspect, several studies have evidenced a high seroprevalence of H pylori among cirrhotic subjects[24-26], but these data have not been ubiquitarily confirmed[27]. Part of these differences might be explained by geographical and clinical differences in the population studied and the study-design[23]. Experimentally, Ward et al[10] have fulfilled Koch’s postulates as far as the association of H. hepaticus with chronic active hepatitis and HCC in mice was concerned. Whether Helicobacter spp. could act as a cofactor in the progression towards cirrhosis and carcinogenesis in humans with viral hepatitis, is still under review[23].
Chronic hepatitis is an inflammatory disease and each inflammatory process is characterised by increased levels of pro-inflammatory cytokines such as interleukins 1, 6 (IL-1, IL-6), tumor necrosis factor (TNF) and by the presence of lympho-mono cellular infiltrate and lymphoid follicle formation[28]. Viruses, such as HCV, are only capable of limited inflammation, due to shedding IL-1 receptor in circulation, thereby limiting the possibility of IL-1 binding to cellular receptors[23]. Helicobacters, on the other hand, are strong inducers of the inflammatory cascade[29], infection with them could lead to the accumulation of extraordinary number of lymphocytes and polymorphonuclear cells in the infected tissue. IL-1 gene cluster polymorphisms, thought to enhance IL-1β production, confer an increased risk of cancer in patients infected with H pylori[30]. Furthermore, Meyer-ter-Vehn et al have recently shown that strains of Cag-A-expressing H pylori could activate the ERK/MAP kinase cascade, resulting in Elk-1 phosphorylation and increased c-fos transcription. Proto-oncogene activation might represent a crucial step in the pathomechanism of H pylori-induced neoplasia. It has been shown that several Helicobacter spp. could also secrete a liver-specific toxin that causes hepatocyte necrosis in cell culture, and might therefore also be involved in damaging liver parenchyma in vivo.
The possibility that Helicobacter spp. could infect the biliary tract and the liver of humans has been reported by several studies in different settings. Fox et al[19] have shown the presence of Helicobacter spp. in the bile of Chileans with chronic cholecystitis. Avenaud and coworkers have demonstrated by PCR the presence of genomic sequences of Helicobacter spp. in the liver of 8 patients with HCC without primary diagnosis of cirrhosis, a further analysis by sequencing revealed that these species were H pylori and Helicobacter felis[18]. Similarly, we have reported the presence of the cagA gene sequences obtained from the liver tissue of cirrhotic patients with HCC. Furthermore, Nilsson et al[17] have identified H pylori and Helicobacter spp. in human liver samples from patients suffering from primary sclerosing cholangitis and primary biliary cirrhosis[17] and recently in liver samples of patients with cholangiocarcinomas or HCC.
Of great interest is the fact that the gene sequence obtained from positive Helicobacter spp. specific 16S rRNA PCR was usually most analogous to H pylori. This encourages the speculation that the presence of Helicobacter DNA in human liver tissue might reflect the transport of H pylori of gastric origin or its DNA to the liver. Studies have also indicated that intestinal Helicobacter might be implicated in hepatobiliary disease. Recently, for the first time, the culture of a Helicobacter strain from the human liver has been described. So far, few studies have been reported regarding the presence of Helicobacter spp. genomes in liver tissues from patients not suffering from primary chronic liver diseases.
We now report the infrequent presence of genomic sequences of H pylori in liver parenchyma of patients operated upon for metastasis to the liver arising from colon cancer. In contrast, the presence of genome of Helicobacter spp. was found with a higher frequency in the liver from patients with HCV-related cirrhosis and HCC. This finding lends support to the hypothesis that co-infection with H pylori or Helicobacter spp. might amplify the chronic inflammation of the parenchyma, thereby leading to cirrhosis and HCC[23]. To date, we do not know whether bacterial infection of the liver could play a pathogenic role in the development of human HCC, analogous to what has been observed in the mouse model, and the question awaits clarification.
We thank Dr. J. Stanley for providing Helicobacter pullorum DNA.
Edited by Wang XL Proofread by Zhu LH
| 1. | Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer. 1999;80:827-841. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 20] [Reference Citation Analysis (0)] |
| 2. | Sherlock S, Dooley J. Diseases of the liver and biliary system. 10th Edition.. Oxford: Blackwell Scientific Publications. 1997;. [Cited in This Article: ] |
| 3. | Maio G, d'Argenio P, Stroffolini T, Bozza A, Sacco L, Tosti ME, Intorcia M, Fossi E, d'Alessio G, Kondili LA. Hepatitis C virus infection and alanine transaminase levels in the general population: a survey in a southern Italian town. J Hepatol. 2000;33:116-120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 101] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 4. | Guadagnino V, Stroffolini T, Rapicetta M, Costantino A, Kondili LA, Menniti-Ippolito F, Caroleo B, Costa C, Griffo G, Loiacono L. Prevalence, risk factors, and genotype distribution of hepatitis C virus infection in the general population: a community-based survey in southern Italy. Hepatology. 1997;26:1006-1011. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 249] [Cited by in F6Publishing: 264] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 5. | Silini E, Bottelli R, Asti M, Bruno S, Candusso ME, Brambilla S, Bono F, Iamoni G, Tinelli C, Mondelli MU. Hepatitis C virus genotypes and risk of hepatocellular carcinoma in cirrhosis: a case-control study. Gastroenterology. 1996;111:199-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 134] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 6. | Wiese M, Berr F, Lafrenz M, Porst H, Oesen U. Low frequency of cirrhosis in a hepatitis C (genotype 1b) single-source outbreak in germany: a 20-year multicenter study. Hepatology. 2000;32:91-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 249] [Cited by in F6Publishing: 257] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 7. | Peano G, Menardi G, Ponzetto A, Fenoglio LM. HLA-DR5 antigen. A genetic factor influencing the outcome of hepatitis C virus infection. Arch Intern Med. 1994;154:2733-2736. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 70] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Cassell GH. Infectious causes of chronic inflammatory diseases and cancer. Emerg Infect Dis. 1998;4:475-487. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 50] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 9. | Mosnier JF, Scoazec JY, Marcellin P, Degott C, Benhamou JP, Feldmann G. Expression of cytokine-dependent immune adhesion molecules by hepatocytes and bile duct cells in chronic hepatitis C. Gastroenterology. 1994;107:1457-1468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 43] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 10. | Ward JM, Fox JG, Anver MR, Haines DC, George CV, Collins MJ, Gorelick PL, Nagashima K, Gonda MA, Gilden RV. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst. 1994;86:1222-1227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 299] [Cited by in F6Publishing: 272] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 11. | Bulent K, Murat A, Esin A, Fatih K, MMMurat H, Hakan H, Melih K, Mehmet A, Bulent Y, Fatih H. Association of CagA and VacA presence with ulcer and non-ulcer dyspepsia in a Turkish population. World J Gastroenterol. 2003;9:1580-1583. [PubMed] [Cited in This Article: ] |
| 12. | Palmas F, Pellicano R, Massimetti E, Berrutti M, Fagoonee S, Rizzetto M. Eradication of Helicobacter pylori infection with proton pump inhibitor-based triple therapy. A randomised study. Panminerva Med. 2002;44:145-147. [PubMed] [Cited in This Article: ] |
| 13. | Testino G, Cornaggia M, De Iaco F. Helicobacter pylori influence on gastric acid secretion in duodenal ulcer patients diagnosed for the first time. Panminerva Med. 2002;44:19-22. [PubMed] [Cited in This Article: ] |
| 14. | Li S, Lu AP, Zhang L, Li YD. Anti-Helicobacter pylori immunoglobulin G (IgG) and IgA antibody responses and the value of clinical presentations in diagnosis of H. pylori infection in patients with precancerous lesions. World J Gastroenterol. 2003;9:755-758. [PubMed] [Cited in This Article: ] |
| 15. | Roussos A, Philippou N, Gourgoulianis KI. Helicobacter pylori infection and respiratory diseases: a review. World J Gastroenterol. 2003;9:5-8. [PubMed] [Cited in This Article: ] |
| 16. | Yakoob J, Jafri W, Abid S. Helicobacter pylori infection and micronutrient deficiencies. World J Gastroenterol. 2003;9:2137-2139. [PubMed] [Cited in This Article: ] |
| 17. | Nilsson HO, Taneera J, Castedal M, Glatz E, Olsson R, Wadström T. Identification of Helicobacter pylori and other Helicobacter species by PCR, hybridization, and partial DNA sequencing in human liver samples from patients with primary sclerosing cholangitis or primary biliary cirrhosis. J Clin Microbiol. 2000;38:1072-1076. [PubMed] [Cited in This Article: ] |
| 18. | Avenaud P, Marais A, Monteiro L, Le Bail B, Bioulac Sage P, Balabaud C, Mégraud F. Detection of Helicobacter species in the liver of patients with and without primary liver carcinoma. Cancer. 2000;89:1431-1439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 10] [Reference Citation Analysis (0)] |
| 19. | Ponzetto A, Pellicano R, Leone N, Cutufia MA, Turrini F, Grigioni WF, D'Errico A, Mortimer P, Rizzetto M, Silengo L. Helicobacter infection and cirrhosis in hepatitis C virus carriage: is it an innocent bystander or a troublemaker. Med Hypotheses. 2000;54:275-277. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 60] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 20. | Ishak KG, Anthony PP, Sobin LH. Histological typing of tumors of the liver. World Health Organization. New York:. Springer Verlag. 1994;. [DOI] [Cited in This Article: ] |
| 21. | Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791-5795. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 883] [Cited by in F6Publishing: 919] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 22. | Sun HC, Tang ZY. Preventive treatments for recurrence after curative resection of hepatocellular carcinoma--a literature review of randomized control trials. World J Gastroenterol. 2003;9:635-640. [PubMed] [Cited in This Article: ] |
| 23. | Fagoonee S, Pellicano R, Rizzetto M, Ponzetto A. The journey from hepatitis to hepatocellular carcinoma. Bridging role of Helicobacter species. Panminerva Med. 2001;43:279-282. [PubMed] [Cited in This Article: ] |
| 24. | Siringo S, Vaira D, Menegatti M, Piscaglia F, Sofia S, Gaetani M, Miglioli M, Corinaldesi R, Bolondi L. High prevalence of Helicobacter pylori in liver cirrhosis: relationship with clinical and endoscopic features and the risk of peptic ulcer. Dig Dis Sci. 1997;42:2024-2030. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 59] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Spinzi G, Pellicano R, Minoli G, Terreni N, Cutufia MA, Fagoonee S, Rizzetto M, Ponzetto A. Helicobacter pylori seroprevalence in hepatitis C virus positive patients with cirrhosis. The Como cross-sectional study. Panminerva Med. 2001;43:85-87. [PubMed] [Cited in This Article: ] |
| 26. | Pellicano R, Leone N, Berrutti M, Cutufia MA, Fiorentino M, Rizzetto M, Ponzetto A. Helicobacter pylori seroprevalence in hepatitis C virus positive patients with cirrhosis. J Hepatol. 2000;33:648-650. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 27. | Chen JJ, Changchien CS, Tai DI, Chiou SS, Lee CM, Kuo CH. Role of Helicobacter pylori in cirrhotic patients with peptic ulcer. A serological study. Dig Dis Sci. 1994;39:1565-1568. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 33] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow. Lancet. 2001;357:539-545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5245] [Cited by in F6Publishing: 5425] [Article Influence: 235.9] [Reference Citation Analysis (0)] |
| 29. | Crabtree J. Cytokine responses to Helicobacter pylori-induced infection. In: Riecken EO, Zeitz M, Stallmach A, Heise W editors. Malignancy and chronic inflammation in the gastro-intestinal tract: new concepts. Lancaster: Kluwer Academic Publishers. 1995;25-36. [Cited in This Article: ] |
| 30. | El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398-402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1690] [Cited by in F6Publishing: 1614] [Article Influence: 67.3] [Reference Citation Analysis (0)] |